Abstract
Purpose: The study purpose was to examine the consequences of using dried blood spots rather than fresh sera in first-trimester Down syndrome screening.
Methods: We collected and compared human chorionic gonadotropin and pregnancy-associated plasma protein-A results from clients providing dried blood spots (Cohort 1) and from other clients providing fresh sera (Cohort 2). Inclusion and exclusion criteria aimed at ensuring the two cohorts were similar.
Results: The average concentrations of human chorionic gonadotropin and pregnancy-associated plasma protein-A are significantly different for the two cohorts. When the results are converted to multiples of the median and weight-adjusted, the variances for human chorionic gonadotropin and pregnancy-associated plasma protein-A in Cohort 1 are greater by 25% and 14%, respectively. Modeling the impact of this increased variance shows that Down syndrome detection is expected to be lower in Cohort 1 (83% vs. 85% at a 5% false-positive rate) or the false-positive rate is expected to be higher (3.9% vs. 3.0% at an 80% detection rate).
Conclusions: This study of two closely matched cohorts provides indirect evidence that dried blood spots will result in slightly lower Down syndrome screening performance. Studies should be undertaken to confirm and further quantify differences in assigned risks by a direct comparison using matched serum and dried blood spots collected from the same women.
Similar content being viewed by others
Main
Virtually all prenatal screening protocols for Down syndrome use measurements made in maternal serum, sometimes with the inclusion of first-trimester ultrasound determination of nuchal translucency (NT) thickness. In a few reports, these serum markers have been measured by using dried blood spots rather than fresh sera.1–4 Collection of blood spots is less invasive than drawing blood and is sometimes more convenient for office personnel and the patient because referral to a phlebotomy station is not required. Also, some analytes can be more stable in blood spots than in whole blood or serum when shipping and handling occurs under conditions such as elevated temperature (e.g., the free β-subunit of human chorionic gonadotropin [hCG]).5 There is limited information concerning possible differences introduced by the choice of sample type. To our knowledge, no study has made a direct (matched) comparison of the screening performance of maternal Down syndrome markers using measurements obtained from a dried blood spot and maternal serum collected from the same women. Matched studies have been undertaken for other analytes, such as newborn screening markers6 and prostate-specific antigen.7 Although not as powerful as a matched study, a cohort study design can yield information on the relative performance of the two sample types when results are compared from women screened who provided blood spots with those screened who provided maternal serum. The current study was undertaken to determine whether any meaningful differences attributable to collection method are observed between two similar cohorts of women being tested for hCG and pregnancy-associated plasma protein-A (PAPP-A). Samples were collected as part of the routine clinical service at Genzyme Genetics.
METHODS
Blood collection
The method used for collecting blood from finger sticks was published in National Committee on Clinical Laboratory Standards document LA4-A2 1992.8 Many of the centers that provided dried blood spots had already been trained for finger stick collection. When a new client is registered, a representative from Genzyme Genetics visits the client to provide information and training on blood spot collection, along with a written protocol. Both serum and dried blood spots were sent by FedEx for next-day delivery. Internal studies showed that both specimen types are stable for 7 days at room temperature; samples older than 7 days are rejected. Exclusion criteria included insufficient volume to fill a circle, spots of irregular shape, spots that were not impregnated throughout, spots with multiple spots within the circle, spots not thoroughly dried, spots contaminated with foreign substances, or evidence of blood clots.
Assay descriptions: Dried blood spots
Both the hCG and PAPP-A immunoassays used to assay dried blood spots are enzymatically amplified “two-step” sandwich-type immunoassays. Calibrators and controls provided with each kit were in liquid form. The intact hCG assay was performed using the ActiveGLO Dried Blood Spot I-hCG Enzyme-Linked Immunosorbent Assay (Diagnostic Systems Laboratories, Webster, TX), following the instructions provided in the package insert. Analytic sensitivity is 0.23 mIU/mL. The PAPP-A assay was performed using the ActiveGLO Dried Blood Spot PAPP-A Enzyme-Linked Immunosorbent Assay (Diagnostic Systems Laboratories), following the instructions provided in the package insert. The analytic sensitivity is 0.21 μIU/mL. Patient values in μIU/mL were multiplied by 250, as described in the package insert, to yield serum-equivalent units of mIU/mL. For hCG, the coefficients of variation for liquid controls provided by the manufacturer, with average values of 34.8 and 319.7 mIU/mL, were 5.0% and 4.1%, respectively, over 18 assay runs; for PAPP-A the coefficients of variation for liquid controls provided by the manufacturer, with average values of 4.9 and 15.4 μIU/mL, were 3.6% and 3.3%, respectively, over 18 assay runs. An in-house blood spot control, made using whole blood collected from a woman in the first trimester, yielded coefficients of variation of 7.4% and 11.5% for hCG and PAPP-A, respectively,
Assay descriptions: Fresh serum
The PAPP-A assay was performed on the DPC Immulite 2000 (Diagnostic Products Corporation, Los Angeles, CA). The Immulite 2000 PAPP-A is a solid-phase, enzyme-labeled chemiluminescent immunometric assay. Analytic sensitivity is 25 μIU/mL. Values of 10 mIU/mL are routinely diluted. The Immulite 2000 total hCG is a solid-phase, two-site chemiluminescent immunometric assay run at a 1:100 on-board dilution. Analytic sensitivity is 0.4 mIU/mL. For hCG, the coefficients of variation for liquid controls supplied by the manufacturer were 5.3%, 4.9%, and 5.1%, with average values of 13.1, 38.2, and 75.3 mIU/mL for 180 to 183 data points, respectively; for PAPP-A the coefficients of variation for liquid controls supplied by the manufacturer were 5.4% and 4.7% with average values of 0.43 and 2.19 mIU/mL for 283 to 287 data points, respectively. An in-house control yielded coefficients of variations of 5.8% and 5.1% for hCG and PAPP-A, respectively, for the same number of data points given above. Rejection criteria for serum samples included hemolysis and severe lipemia.
Selection of the cohorts
Assay results from consecutive samples received over a 1-month period in 2006 were retrieved from clinical patient records. Inclusion criteria included white mother with ultrasound-determined gestational age between 10 weeks, 3 days and 13 weeks, 6 days, with both maternal age and weight available. The data were then de-identified at the laboratory site and sent electronically to Women and Infants Hospital for analysis. The study was approved by the institutional review board at Women and Infants Hospital of Rhode Island.
Modeling Down syndrome screening performance
Assay results were converted to weight-adjusted multiples of the median (MoM). Expected Down syndrome screening performance (detection and associated false-positive rates) was modeled using overlapping multivariate Gaussian distributions, as described earlier.9 The parameters describing these distributions were taken from the literature,10 but the standard deviations and correlation coefficients were modified, on the basis of the distribution of unaffected pregnancies observed in the present study, using a published methodology.11 These modified parameters allow the calculation of likelihood ratios specific to the characteristics of dried blood spots and fresh sera. These likelihood ratios were then multiplied by the age-associated risk to generate an individual Down syndrome risk. At selected risk cutoff levels, it is possible to compute an expected Down syndrome detection and false-positive rate. We used the maternal age distribution for the United States in 2000,12 where the median age is 27 years and 13% of the women are aged 35 years or more. Maternal age-associated risks for Down syndrome13 and assumptions of a 43% loss of fetuses affected with Down syndrome between the late first trimester and term14 were used to assign first-trimester risks. Approximate 95% confidence intervals around the differences in detection (or false-positive rates) were generated using a Monte Carlo method described in an earlier publication.15 Briefly, a simulation was performed using randomly selected logarithmic means, standard deviations, and correlation coefficients for hCG and PAPP-A measurements in fresh sera and again in dried blood spots for both cases and controls. The random selection was from a Gaussian distribution centered on the reported values with a standard error derived from the reported standard deviations and numbers of samples. The corresponding parameters for NT remained constant for each simulation. The resulting performance estimates for 40 of these simulations were sorted, and the observed confidence range was reported.
RESULTS
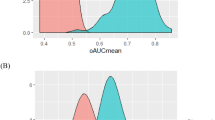
Table 1 shows demographic and biochemical test results for the two cohorts. Overall, the maternal age, weight, and race/ethnicity were nearly identical. The gestational age distributions were similar, except for a small excess of samples at 10 weeks' gestation for the serum samples. The hCG measurements in dried blood spots were significantly lower at all weeks (P < 0.001, analysis of variance after logarithm transformation) compared with hCG measurements in fresh serum samples. PAPP-A measurements were also significantly different for the two sample types, but they were higher rather than lower with dried blood spots (P < 0.001). Separate median values were then computed for each analyte for each cohort, and the results were converted to MoM. The MoM levels were then adjusted for maternal weight according to a reciprocal model,16 with coefficients derived from each cohort for each analyte. The resulting weight-adjusted MoM levels for hCG and PAPP-A were then plotted on a probability plot to determine how well they fit a Gaussian distribution (Figs. 1 and 2).
These data were then used to derive the logarithmic mean and standard deviation by regressing the values within the linear region of the probability plot (5th–95th centiles). All four sets of data fitted a logarithmic Gaussian distribution well. The median values were all 1.00 (logarithmic mean of 0.000) with standard deviations for hCG of 0.1985 and 0.1778 for dried blood spots and fresh sera. The higher variance (+25%) for hCG in the dried blood spot cohort is significantly higher (F = 1.25, P = 0.01) than in the fresh sera cohort. The standard deviations for PAPP-A are 0.2637 and 0.2467 for dried blood spots and fresh sera, respectively. The 14% increase in variance using dried blood spots for PAPP-A is not statistically significant (F = 1.14, P = 0.14).
To estimate the impact on screening performance of any increased variability in the cohort providing dried blood spots versus fresh sera, we used a published method of modifying consensus screening parameters at 12 weeks' gestation10 to parameters that are specifically tailored to each sample type. This methodology is based on the observed variance of measurements in the unaffected population.11 Parameters for NT measurements are also taken from the Serum, Urine and Ultrasound Screening Study (SURUSS) study (logarithmic mean, standard deviation of 0.2922, 0.2313 and 0.000, 0.1329 in Down syndrome and unaffected pregnancies, respectively). Correlations between the NT and biochemical markers are taken to be zero. Truncation limits are those specified in SURUSS. Table 2 summarizes the biochemical parameters used in modeling.
The results of the modeling are shown in Table 3. Overall, the expected Down syndrome screening performance is lower, using the parameters derived from the dried blood spot cohort. At a 5% false-positive rate, the detection rate is 83% versus 85%. The approximate 95% confidence interval around the difference of 3% is from −3% to 9% (not statistically significant). At an 80% detection rate, the false-positive rate is 3.9% versus 2.9%. Most of this variability derives from the original dataset from which the parameters are taken (SURUSS) and will not be reduced with greater numbers of observations in unaffected pregnancies. Overall, 0.2% of dried blood spots were rejected as unsuitable for assay because of insufficient sample (quantity not sufficient); none were rejected on the basis of assay results. For comparison, 0.3% of serum samples were rejected (0.2% hemolyzed, 0.1% quantity not sufficient); none were rejected on the basis of assay results.
DISCUSSION
The first application of dried human blood spots collected on filter paper was for the measurement of phenylalanine for the detection of phenylketonuria in newborns.17 Since that time, dried blood spots have been routinely used in newborn screening for a variety of metabolic diseases. Dried blood spots offer the potential advantages of ease of collection, transport, and storage, as well as possibly reducing the degradation of analytes attributable to extremes of temperature or transport delays. The risk of transmission of bloodborne pathogens may also be reduced. Given these advantages, researchers have investigated the possibility of using blood spots as a collection method for prenatal screening. An early report showed that α-fetoprotein could be reliably measured using dried blood spots for the detection of fetal neural tube defects.18 More recently, immunofluorometric α-fetoprotein and intact hCG assays have been adapted for measurement of these analytes in dried blood spots for the purposes of prenatal screening for neural tube defects and Down syndrome.19 Once the feasibility of measurement of prenatal screening markers using dried blood spots has been demonstrated, the next step is to conduct intervention studies to gain information on screening performance in practice. A number of these have been published, and in general the screening performance using dried blood spots has been comparable to that reported for serum.1,2,4 However, these reports did not directly compare the results obtained using blood spots with those obtained using serum, but instead compared their screening performance (detection rate and false-positive rate) with other published studies that used serum. This approach has the limitation that factors other than sample type will influence these estimates. The most powerful method for comparing the relative screening performance of measurements made using blood spots with those using serum would be a study design in which each woman provided both sample types. Such a matched analysis would control for all of the variables that influence the final estimate of screening performance (e.g., method of gestational dating, risk cutoff used, the maternal age distribution of the screened population, and parameters used to assign risk).
Large matched studies might be difficult, because each woman would need to agree to have both a blood spot and serum collected. Women being screened using the blood spot collection method might be reluctant to provide a serum sample for research purposes, because the blood spot protocol is promoted as being less invasive. Matched studies, however, have been successfully performed for other analytes such as newborn screening markers6 and prostate-specific antigen.7
Although not as powerful as a matched study, information on the relative performance of the two methods of sample collection can be gained by comparing results obtained from a cohort of women screened who provided blood spots with a cohort of women screened who provided serum in which many of the confounding variables (e.g., differing maternal ages and risk algorithms) have been removed. This was the design of the present study, and it provides baseline data needed to perform a robust comparison of Down syndrome screening performance. It relies on estimates of the variance of hCG and PAPP-A MoM levels in unaffected pregnancies for the two sample types. The variances of both the PAPP-A and the hCG measurements were greater for blood spots compared with serum, by 14% and 25%, respectively. Modeling the screening performance of the two serum markers with NT and maternal age yields an estimated 83% detection rate at a 5% false-positive rate for blood spots compared with an 85% detection rate for serum or a 3.9% false-positive rate for blood spots versus 3.0% for serum at an 80% detection rate. These differences are small enough that they are unlikely to be noticeable by screening programs during practice.
The increased variance observed for analytes measured using blood spots is not unexpected, because there are additional sources of variability that can influence these measurements (e.g., lot-to-lot differences in filter paper, between-patient differences in hematocrit, and imprecision in the volume contained in the blood spot punch20). The quality of the blood spot may also vary, depending on the experience and the diligence of the collection personnel. Blood spots collected by patients would be expected to introduce another source of variability, because the patients are inexperienced in sample collection. Our observed increase in variability for PAPP-A and hCG results in dried blood spots may not be applicable to other analytes. For example, one study found that free β-hCG measurements were less variable using blood spots than those reported in other studies using maternal serum.2 However, it is known that free β-concentrations increase in maternal serum on storage or with elevated temperatures, and the use of blood spots may prevent this increase.21 Finding reduced variance for other markers in blood spots compared with fresh sera would not, therefore, be expected for other Down syndrome markers, all of which are stable in maternal serum.22
Estimation of detection and false-positive rates for any given screening marker protocol (efficiency) using modeling sets the upper limit of expected performance. Two protocols with identical screening performance by modeling may differ in practice because one of the protocols has implementation issues. For example, the overall effectiveness of screening protocols that use NT measurements will be less than projected if a certain percentage of those measurements cannot be obtained. This was the case for 7% of the women in the First and Second Trimester Evaluation of Risk of Aneuploidy study.23 Along the same lines, blood spots might be expected to be less effective than serum protocols. Some blood spots would be rejected on receipt because they do not meet the criteria established as essential for obtaining reliable results (e.g., circles are not completely filled, samples were not completely dry when sent, clots were visible on the surface of the sample).
The current study provides evidence that use of blood spots as a sample type may be somewhat less efficient than use of serum when measuring PAPP-A and hCG. More definitive information on the relative merits of the two sample types could be obtained by a matched analysis in a blood spot and serum sample obtained from each woman. A properly designed study would also allow an evaluation of the quality of blood spots obtained from different collection sites. However, the overall differences in performance can be expected to be small, and the use of blood spots as a sample type may be justified in certain situations.
References
Krantz DA, Hallahan TW, Orlandi F, Buchanan P, et al. First-trimester Down syndrome screening using dried blood biochemistry and nuchal translucency. Obstet Gynecol 2000; 96: 207–213.
Macri JN, Anderson RW, Krantz DA, Larsen JW, et al. Prenatal maternal dried blood screening with alpha-fetoprotein and free beta-human chorionic gonadotropin for open neural tube defect and Down syndrome. Am J Obstet Gynecol 1996; 174: 566–572.
Orlandi F, Damiani G, Hallahan TW, Krantz DA, et al. First-trimester screening for fetal aneuploidy: biochemistry and nuchal translucency. Ultrasound Obstet Gynecol 1997; 10: 381–386.
Verloes A, Schoos R, Herens C, Vintens A, et al. A prenatal trisomy 21 screening program using alpha-fetoprotein, human chorionic gonadotropin, and free estriol assays on maternal dried blood. Am J Obstet Gynecol 1995; 172: 167–174.
Spencer K, Macri JN, Carpenter P, Anderson R, et al. Stability of intact chorionic gonadotropin (hCG) in serum, liquid whole blood, and dried whole-blood filter-paper spots: impact on screening for Down syndrome by measurement of free beta-hCG subunit. Clin Chem 1993; 39: 1064–1068.
Hannon WH, Baily CM, Bartoshesky LE, Davin B, et al. Blood collection of filter paper for newborn screening programs, approved standards. National Committee for Clinical Laboratory Standards Document–LA4-A4. Fourth edition. Wayne, PA: National Committee for Clinical Laboratory Standards, 2003, 1–40.
Hoffman BR, Yu H, Diamandis EP . Assay of prostate-specific antigen from whole blood spotted on filter paper and application to prostate cancer screening. Clin Chem 1996; 42: 536–544.
National Committee for Clinical Laboratory Standards, Blood collection on filter paper for neonatal programs. 2nd edition: approved standard. Villanova, PA: National Committee for Clinical Laboratory Standards Publications LA4-A2, 1992.
Royston P, Thompson SG . Model-based screening by risk with application to Down's syndrome. Stat Med 1992; 11: 257–268.
Wald NJ, Rodeck C, Hackshaw AK, Walters J, et al. First and second trimester antenatal screening for Down's syndrome: the results of the Serum, Urine and Ultrasound Screening Study (SURUSS). J Med Screen 2003; 10: 56–104.
Cuckle H . Improved parameters for risk estimation in Down's syndrome screening. Prenat Diagn 1995; 15: 1057–1065.
Centers for Disease Control. Vital and Health Statistics 2000-Natality Data Set, Series 21, No. 14 [Database on CD-ROM]. Hyattsville, MD: Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Health Statistics, 2002;
Hecht CA, Hook EB . Rates of Down syndrome at livebirth by one-year maternal age intervals in studies with apparent close to complete ascertainment in populations of European origin: a proposed revised rate schedule for use in genetic and prenatal screening. Am J Med Genet 1996; 62: 376–385.
Morris JK, Wald NJ, Watt HC . Fetal loss in Down syndrome pregnancies. Prenat Diagn 1999; 19: 142–145.
Canick JA, Lambert-Messerlian GM, Palomaki GE, Neveux LM, et al. Comparison of serum markers in first-trimester down syndrome screening. Obstet Gynecol 2006; 108: 1192–1199.
Neveux LM, Palomaki GE, Larrivee DA, Knight GJ, et al. Refinements in managing maternal weight adjustment for interpreting prenatal screening results. Prenat Diagn 1996; 16: 1115–1119.
Guthrie R, Susi A . A simple phenylalanine method for detecting phenylketonuria in large populations of newborn infants. Pediatrics 1963; 32: 338–343.
Wong PY, Mee AV, Doran TA . Studies of an alpha-fetoprotein assay using dry blood-spot samples to be used for the detection of fetal neural tube defects. Clin Biochem 1982; 15: 170–172.
Vieira Neto E, Carvalho EC, Fonseca A . Adaptation of alpha-fetoprotein and intact human chorionic gonadotropin fluoroimmunometric assays to dried blood spots. Clin Chim Acta 2005; 360: 151–159.
Mei JV, Alexander JR, Adam BW, Hannon WH . Use of filter paper for the collection and analysis of human whole blood specimens. J Nutr 2001; 131: 1631S–1636S.
Beaman JM, Akhtar N, Goldie DJ . Down's syndrome screening using free beta hCG: instability can significantly increase the Down's risk estimate. Ann Clin Biochem 1996; 33 ( Pt 6): 525–529.
Lambert-Messerlian GM, Eklund EE, Malone FD, Palomaki GE, et al. Stability of first- and second-trimester serum markers after storage and shipment. Prenat Diagn 2006; 26: 17–21.
Malone FD, Canick JA, Ball RH, Nyberg DA, et al. First-trimester or second-trimester screening, or both, for Down's syndrome. N Engl J Med 2005; 353: 2001–2011.
Acknowledgements
We thank the American College of Medical Genetics Foundation for partial support of this project.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure: All authors are associated with programs providing prenatal screening for Down syndrome. Genzyme Genetics (J. L.) offers prenatal screening using dried blood spots. No other conflicts of interest exist.
Rights and permissions
About this article
Cite this article
Palomaki, G., Neveux, L., Knight, G. et al. Estimating first-trimester combined screening performance for Down syndrome in dried blood spots versus fresh sera. Genet Med 9, 458–463 (2007). https://doi.org/10.1097/GIM.0b013e31809861a9
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1097/GIM.0b013e31809861a9