Abstract
Purpose: Nephrogenic Diabetes Insipidus (NDI) is genetically heterogeneous and may be inherited in an X-linked or autosomal recessive manner. We aimed to investigate the molecular basis of NDI among Arab families.
Methods: Direct sequencing of coding regions for AQP2 and AVPR2 was used to identify underlying mutations. One large deletion required Southern blot analysis and a PCR-based strategy to identify deletion junctions.
Results: We identified two novel missense mutations (AQP2:p.Gly100Arg and p.Gly180Ser) in AQP2 and one novel missense mutation (AVPR2:p.Gly122Asp), one previously reported missense mutation (AVPR2:p.Arg137His) and one novel contiguous deletion (AVPR2:c.25 + 273_ARHGAP4o:2650-420del) affecting AVPR2. We also describe evidence of lyonization associated with the novel deletion.
Conclusions: Two novel mutations were identified in each of AVPR2 and AQP2 underlying CNDI in Arab families. Identification of these mutations will facilitate early diagnosis of CNDI, counseling of families and provide opportunities for early intervention aimed at reducing morbidity. The presence of affected females and consanguinity, as is often observed in Arab communities should not be used to rule out AVPR2 as a candidate when considering diagnostic testing. Careful observation of phenotypic heterogeneity should be used in referring such families for both AQP2 and AVPR2 molecular genetic testing.
Similar content being viewed by others
Main
AVPR2 and AQP2, the genes coding for the vasopressin type 2 receptor and the aquaporin-2 water channel protein, respectively, have been shown to be critical in the complex system of normal fluid regulation in the body.1,2 Congenital nephrogenic diabetes insipidus (CNDI) is an inherited disorder, caused by mutations in AVPR2 or AQP2.3–7 NDI patients are unable to increase urinary osmolality, which remains <200 mosmol/kg H2O (normal values are >805 mosmol/kg H2O). CNDI may be inherited in either X-linked (MIM# 304800) or autosomal recessive (MIM# 125800) modes of inheritance. AVPR2 encodes the arginine vasopressin receptor type II (AVPR2) and is located on the X-chromosome at Xq28. Therefore, AVPR2 related CNDI exhibits an X-linked inheritance pattern. AVPR2 is a 371 amino acid protein with 7 transmembrane-spanning domains and is a member of the G protein-coupled receptor (GPCR) superfamily.8 AQP2 is the gene coding for the aquaporin-2 water channel protein (AQP2) and is located on chromosome 12 at 12q13.9 The aquaporin-2 water channel protein is 271 amino acids long, contains 6 membrane-spanning regions and is a member of the membrane integral protein (MIP)/water channel family. It is expressed exclusively in collecting duct principal cells and medullary collecting duct cells, the cells in which vasopressin-regulated water resorption takes place.10 Mutations in AQP2 have been reported in 10% of cases of CNDI.11 The inheritance pattern is usually autosomal recessive, however dominant inheritance has been reported, albeit rarely.12
Arab populations in which consanguinity and endogamy is common remain relatively culturally and geographically isolated13 and largely understudied as evidenced by the finding of novel, generally population-specific mutations, even for common, well-studied disorders such as cystic fibrosis.14 Despite 178 AVPR2 and 34 AQP2 mutations and being listed by the Human Gene Mutation Database (HGMD),15 none and only 2, respectively, have been associated with Arabs to date, highlighting the need for this study.
MATERIALS AND METHODS
Study subjects
Four families from the Pediatric Nephrology clinics at King Faisal Specialist Hospital and Research Centre (KFSHRC) and one family from the Security Forces Hospital (SFH) in Riyadh were used for this study. Family 1 was part of an Institutional Review Board approved research project and all individuals were fully consented. Families 2–5 were referred for diagnostic testing. In all instances data has been anonymized for publication. The water deprivation test and renal manifestations such as urinary output following DDAVP administration was part of the standard diagnostic workup in all patients. However, no additional clinical records were available to differentiate patients with AVPR2 or AQP2 related pathogenesis. Clinical status was based primarily upon the level of polyuria with consideration of the absence or presence of the following additional symptoms: hyperosmolality, hypernatremia, failure to thrive, renal impairment secondary to dehydration and central nervous system involvement (brain damage, mental retardation). Normal urinary output is 0.5–2.0 mL/kg/hour. For the purposes of this study, mild NDI was characterized by polyuria of 2–4 mL/kg/hour with few other symptoms; polyuria of 4–5 mL/kg/hour was classified as moderate disease; and severe disease was characterized by a urinary output of > 5 mL/kg/hour.
Sample collection and genomic DNA extraction
Genomic DNA was extracted from whole blood anticoagulated with EDTA using the Purgene Gentra DNA Extraction Kit (Cat. # D-5000, Gentra Systems, Minneapolis, MN) according to the manufacturers instructions. The DNA was quantified spectrophotometrically and stored in aliquots at −20°C until required.
PCR amplification and direct sequencing of AVPR2 and AQP2
PCR was performed using previously published primer pairs and conditions to amplify the entire coding region of both AVPR2 16 and AQP2.6 Direct sequencing was performed using the DYEnamic ET Dye Terminator Cycle Sequencing Kit (Cat. # US81060, GE Health Care, CA) for MegaBACE DNA Analysis Systems using the manufacturer's protocol. Samples were analyzed on the MegaBACE 1000 96-capillary sequencer. Sequence data were analyzed using the DNASTAR SeqManII DNA analysis software. Reference sequences used were GenBank #L22206 for AVPR2 and GenBank #AF147092 for AQP2.
Southern blot analysis
Restriction enzyme digestion with Pst I was performed using 8 μg of genomic DNA per sample analyzed. Digestion products were run on a 1% agarose gel in Tris-borate EDTA (TBE) buffer. Blotting to Nylon membrane (Hybond-N+, GE Health Care, CA) was performed by vacuum transfer. Nucleic acid was then fixed to the membrane by UV cross-linking after which the blot was prehybridized for 1 hour at 65°C in 25 mL of 5X SSPE, 5X Denhardt's Solution, 0.5% SDS and 50 μg of sonicated salmon sperm DNA. The probe was prepared by labeling a 345bp PCR product generated by amplification of nucleotides 189 through 534 of AVPR2 with α-32P dCTP using the Ready To Go DNA Labeling Kit (Pharmacia Biotech, Piscataway, NJ). The probe was added to prehybridization solution and the membrane incubated overnight at 65°C. The blot was washed in 2 × SSPE, 0.1% SDS at 65°C for 15 minutes followed by a 1 × SSPE, 0.1% SDS wash at 65°C for 10 minutes, prior to autoradiography.
RESULTS
Families
Family 1: There were three affected male and two unaffected female children in this family. Blood samples were available from both parents and all three affected males. The parents are unaffected and the three male siblings all suffer from moderate disease, which is consistent with an X-linked form of inheritance.
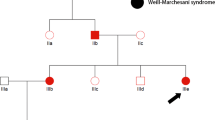
Family 2: This three-generation family was initially thought to demonstrate an autosomal recessive inheritance pattern, as there were two affected females and four affected males. Grandparents were consanguineous and the grandmother was mildly affected. She gave birth to nine children, four male and five female. Three of the four males were also severely affected with NDI. Two of the five females died of unknown causes and one of the remaining three females was moderately affected with NDI and had three children, one moderately affected male and two unaffected females.
Family 3: There was a single sample available from this family, a mildly affected male child.
Family 4: This family consists of two unaffected parents and one affected female child, consistent with an autosomal recessive pattern of inheritance.
Family 5: Blood samples were available from all six members of this consanguineous family: two unaffected parents; three severely affected children (one female and two male) and one unaffected male child. The pedigree was consistent with an autosomal recessive mode of inheritance.
Genotyping
Direct sequencing of the entire coding regions for AQP2 and AVPR2 in five Saudi Arabian families revealed four novel and one previously described mutation. The first, a novel missense mutation found in AQP2 segregated with disease in Family 5 and was a transition, AQP2:c.298G>A (p.Gly100Arg) that substituted arginine for glycine in the third transmembrane region of the protein. The three affected individuals were homoallelic for the mutation with parents and the unaffected male all being carriers. The second, also novel, mutation of AQP2 in Family 4 was another transition, AQP2:c.537G>A (p.Gly180Ser), in exon 3 resulting in the substitution of glycine by serine in the third extracellular loop of AQP2. The one affected individual from this family was homoallelic for the mutation with the parents being heterozygous carriers. One hundred chromosomes from unaffected individuals were surveyed by direct sequencing and found to be free of these AQP2 sequence alterations.
Two missense mutations were found in AVPR2. The first from Family 1 was a novel mutation, AVPR2:c.365G>A (p.Gly122Asp), present in all three affected male children. The mother was an asymptomatic heterozygote and the father had the wildtype G allele. Direct sequencing of one hundred chromosomes from unaffected individuals showed that this variant was absent in the population at large. The AVPR2 mutation found in family III was a transition, AVPR2:c.410G>A (p.Arg137His) in exon 2. The latter mutation present in the single member of this family tested, has previously been reported as underlying CNDI.17
Family 2 harbored a large deletion of AVPR2 (Fig. 1). Attempts to amplify AVPR2 segments from DNA of the affected male members in this family (Fig. 2: II-1, 4 and 9 were unsuccessful with all primer pairs except V2-1 and V2-2 (Fig. 1A). We attempted amplification using V2-1 and V2-3 (internal to V2-1 and V2-2) or V2-6, the most downstream AVPR2 primer (Fig. 1B), however no amplicon was generated using affected male genomic DNA as template. Southern blot analysis of family members revealed the presence of both wildtype and mutant alleles (Fig. 2) and confirmed the presence of a large deletion. The deletion was characterized by amplifying sequential 500 bp fragments, first every 5 kb, for 20 kb, beginning 30 kb downstream of AVPR2 (moving in an upstream direction), and from there every 1–2 kb upstream until the first region that would not amplify in affected males was identified (primer sequences available upon request). We were then able to generate an amplicon containing the deletion breakpoints using primers V2-1 and Xq28-10R. This generated an amplicon of 1246 bp (Fig. 1C). The deletion was characterized by direct sequencing of this fragment (Fig. 1D), which identified a 2932 bp deletion (AVPR2:c.25 + 273_ARHGAP4o:2650-420del) present in all affected males and in the heterozygous state for females II-5 and III-3 (Fig. 2). The consequence to AVPR2 was deletion of the entire gene downstream of the breakpoint located in intron 1 and extending into ARHGAP4 which is transcribed in the opposite orientation to AVPR2. As a result exons 22, 23 and the 3′ untranslated region of ARHGAP4 is also deleted. The immunoglobulin class switch (ICS) motif (5′-TGGGG-3′) was found immediately adjacent to both deletion breakpoints such that one motif was preserved at the deletion junction.
(A) Primers V2-1 and V2-2 amplified a 345 bp fragment in all family members. (B) Primers V2-3 and V2-6 failed to amplify in affected male family members. (C) Primers V2-1 and Xq28-10R cover a region 4178 bp in size, however in affected male family members they amplified a 1246 bp fragment. (D) Solid bars represent the region that was present in affected males and the line indicates the deletion.
Pedigree of Family 2 and corresponding Southern blot. The solid symbols indicate affected individuals, + indicates mild symptoms, ++ moderate symptoms and +++ severe symptoms of NDI. The unaffected male control demonstrates the normal allele; III-1, an affected male, has the allele containing the deletion; II-5, a moderately affected female is heterozygous for both the normal and deletion allele; III-2 is homozygous for the normal allele and III-3 is shown to be a carrier based on Southern blot analysis.
DISCUSSION
Our study is the first to report on the genetic basis of CNDI in an Arab population. Despite 178 AVPR2 mutations being listed by the HGMD, none have been associated with Arabs.15 In this report, we identified the mutation of AVPR2 underlying CNDI in each of three Saudi families. In two other families from our study, CNDI was recessively inherited and two novel mutations [AQP2:c.298G>A (p.Gly100Arg) and AQP2:c.537G>A (p.Gly180Ser)] were identified. Mutations in AQP2 are much less frequent, with the HGMD reporting a total of 34 mutations to date, only two of which were associated with Arabs.15
Our study looked at a total of five families with CNDI. The literature reports that approximately 10% of CNDI cases are attributable to mutations in the AQP2.18 Patients with CNDI due to recessive AQP2 mutations are often compound heterozygotes.19 We report here two families with homozygous mutations in AQP2, which is not surprising given the presence of consanguinity. In Family 5, a transition resulted in a novel missense mutation AQP2:c.298G>A (p.Gly100Arg) altering the third transmembrane domain of the mature protein. Misrouting of AQP2 mutants is the major underlying cause of autosomal recessive CNDI.20 The mutant proteins are not properly routed due to their retention in the quality control component of the endoplasmic reticulum.21 More specific evidence that supports AQP2:c.298G>A (p.Gly100Arg) as the disease causing mutation is provided by other researchers who performed functional expression studies of an AQP2:c.299G>T (p.Gly100Val) mutant in Xenopus oocytes which showed little increase in water permeability when injected with the associated cRNA. In addition, immunocytochemical analysis revealed the absence of the mutant AQP2 in the plasma membrane.19 The HGMD reports the AQP2:c.299G>T (p.Gly100Val) missense mutation and a AQP2:c.298G>T (p.Gly100Stop) nonsense mutation at the same amino acid position.15 The codon changes in these previously reported mutations are GGA to GTA and GGA to TGA, respectively, while the codon change reported here is GGA to AGA. The occurrence of three mutations at this codon of AQP2 may indicate the presence of a mutational hotspot.
The second novel missense AQP2 mutation, identified in Family 4, was another transition, AQP2:c.537G>A (p.Gly180Ser). A survey of more than 100 chromosomes from unrelated, unaffected individuals was negative for this nucleotide base change. Absence of this nonsynonymous allele in the general population and its segregation with disease in a recessive pattern within this family is consistent with it being a pathogenic alteration.
The primary cause of CNDI is mutation of AVPR2. Mutations of AVPR2 are implicated in nearly 90% of all cases of CNDI.22 We identified one novel missense mutation, one previously reported missense mutation and a novel 2932 bp deletion of AVPR2. The novel missense mutation was caused by a transition in exon 2, AVPR2:c.365G>A (p.Gly122Asp) altering the third transmembrane domain of the protein in the middle of a 40 amino acid block (residues 107–147) that is completely conserved across 5 species surveyed (human, mouse, bovine, rat, pig). The literature reports another missense mutation resulting in the substitution of glycine by arginine, AVPR2:c.364G>C (p.Gly122Arg) at this amino acid position of AVPR2,23 further supporting pathogenicity of AVPR2:c.365G>A (p.Gly122Asp).
AVPR2:c.410G>A (p.Arg137His) was previously reported and well characterized.17 This mutation found in the single individual tested from Family 3 is relatively common and was found in eight unrelated families from various geographic locations, i.e., France, Japan, Canada, USA, and Switzerland and now Saudi Arabia.15 This mutation is associated with CpG dinucleotides that are known mutational hot spots for genetic disease.24 The mutant receptor shows normal binding properties, but is unable to stimulate the GS/adenylyl cyclase system.17
The third AVPR2 defect was a novel deletion found in family II. With the combination of consanguinity and affected females, this family appeared to be a classic example of autosomal recessive inheritance and AQP2 was the first candidate screened. When no mutations were found, the pedigree was revisited and the phenotypic heterogeneity of the disease in this family ranging from mild to severe was noted and found to be suggestive of an X-linked pattern of inheritance with Lyonization. When AVPR2 was interrogated, a 2932bp deletion spanning the 3′ regions of both AVPR2 and the adjacent ARHGAP4 was identified. The presence of immunoglobulin class switch (ICS) motifs immediately adjacent to the deletion breakpoints is suggestive of the mutant allele arising from failure of a physiological mechanism such as switch recombination. Indeed a similar mutation process involving the ICS motif was proposed for a previously reported contiguous gene deletion involving AVPR2 and ARHGAP4.25 In addition an extrachromosomal substrate assay for ICS recombination also highlights the central role of the ICS motif in recombination that involves excision of an intervening sequence.26 Interestingly, no symptoms specific to the ARHGAP4 gene were identified in our patients. Contiguous deletions involving AVPR2 and ARHGAP4 have been reported on two previous occasions, with the NDI phenotype not being compounded in either instance.25,27 ARHGAP4 encodes a cytoplasmic protein of 115 kD that is preferentially expressed in hematopoietic cells. One patient with a contiguous deletion of ARHGAP4 and AVPR2 was followed over a 16-year period and included extensive analysis of blood chemical, enzyme and hormone values and showed no deviations from normal ranges other than those related to the CNDI phenotype.27 They postulated that other members of the GTPase-activating protein family most likely compensate the loss of ARHGAP4 function.
In summary, we describe two novel mutations in each of AVPR2 and AQP2 underlying CNDI in Saudi families. Identification of these mutations will facilitate early diagnosis of CNDI, timely clinical intervention and counseling of families. The presence of affected females and consanguinity, as is often observed in Arab communities should not be used to rule out AVPR2 as a candidate when considering diagnostic testing. Careful observation of phenotypic heterogeneity should be used in referring such families for both AQP2 and AVPR2 molecular genetic testing.
References
Fujiwara TM, Bichet DG . Molecular biology of hereditary diabetes insipidus. J Am Soc Nephrol 2005; 16: 2836–2846.
Knoers NV, Deen PM . Molecular and cellular defects in nephrogenic diabetes insipidus. Pediatr Nephrol 2001; 16: 1146–1152.
Pan Y, Metzenberg A, Das S, Jing B, et al. Mutations in the V2 vasopressin receptor gene are associated with X-linked nephrogenic diabetes insipidus. Nat Genet 1992; 2: 103–106.
Rosenthal W, Seibold A, Antaramian A, Gilbert S, et al. Mutations in the vasopressin V2 receptor gene in families with nephrogenic diabetes insipidus and functional expression of the Q-2 mutant. Cell Mol Biol (Noisy-le-grand) 1994; 40: 429–436.
Canfield MC, Tamarappoo BK, Moses AM, Verkman AS, et al. Identification and characterization of aquaporin-2 water channel mutations causing nephrogenic diabetes insipidus with partial vasopressin response. Hum Mol Genet 1997; 6: 1865–1871.
Deen PM, Verdijk MA, Knoers NV, Wieringa B, et al. Requirement of human renal water channel aquaporin-2 for vasopressin-dependent concentration of urine. Science 1994; 264: 92–95.
Hochberg Z, Van Lieburg A, Even L, Brenner B, et al. Autosomal recessive nephrogenic diabetes insipidus caused by an aquaporin-2 mutation. J Clin Endocrinol Metab 1997; 82: 686–689.
Morel F, Imbert-Teboul M, Chabardes D . Receptors to vasopressin and other hormones in the mammalian kidney. Kidney Int 1987; 31: 512–520.
Deen PM, Weghuis DO, Sinke RJ, Geurts van Kessel A, et al. Assignment of the human gene for the water channel of renal collecting duct Aquaporin 2 (AQP2) to chromosome 12 region q12–>q13. Cytogenet Cell Genet 1994; 66: 260–262.
Fushimi K, Sasaki S, Yamamoto T, Hayashi M, et al. Functional characterization and cell immunolocalization of AQP-CD water channel in kidney collecting duct. Am J Physiol 1994; 267: F573–F582.
van Os CH, Deen PM . Aquaporin-2 water channel mutations causing nephrogenic diabetes insipidus. Proc Assoc Am Phys 1998; 110: 395–400.
Kuwahara M, Iwai K, Ooeda T, Igarashi T, et al. Three families with autosomal dominant nephrogenic diabetes insipidus caused by aquaporin-2 mutations in the C-terminus. Am J Hum Genet 2001; 69: 738–748.
Meyer BF . Strategies for the prevention of hereditary diseases in a highly consanguineous population. Ann Hum Biol 2005; 32: 174–179.
Kambouris M, Banjar H, Moggari I, Nazer H, et al. Identification of novel mutations in Arabs with cystic fibrosis and their impact on the cystic fibrosis transmembrane regulator mutation detection rate in Arab populations. Eur J Pediatr 2000; 159: 303–309.
Stenson PD, Ball EV, Mort M, Phillips AD, et al. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat 2003; 21: 577–581.
Pasel K, Schulz A, Timmermann K, Linnemann K, et al. Functional characterization of the molecular defects causing nephrogenic diabetes insipidus in eight families. J Clin Endocrinol Metab 2000; 85: 1703–1710.
Rosenthal W, Antaramian A, Gilbert S, Birnbaumer M . Nephrogenic diabetes insipidus. A V2 vasopressin receptor unable to stimulate adenylyl cyclase. J Biol Chem 1993; 268: 13030–13033.
Boccalandro C, De Mattia F, Guo DC, Xue L, et al. Characterization of an aquaporin-2 water channel gene mutation causing partial nephrogenic diabetes insipidus in a Mexican family: evidence of increased frequency of the mutation in the town of origin. J Am Soc Nephrol 2004; 15: 1223–1231.
Lin SH, Bichet DG, Sasaki S, Kuwahara M, et al. Two novel aquaporin-2 mutations responsible for congenital nephrogenic diabetes insipidus in Chinese families. J Clin Endocrinol Metab 2002; 87: 2694–2700.
Levin MH, Haggie PM, Vetrivel L, Verkman AS . Diffusion in the endoplasmic reticulum of an aquaporin-2 mutant causing human nephrogenic diabetes insipidus. J Biol Chem 2001; 276: 21331–21336.
Kamsteeg EJ, Deen PM, van Os CH . Defective processing and trafficking of water channels in nephrogenic diabetes insipidus. Exp Nephrol 2000; 8: 326–331.
Morello JP, Bichet DG . Nephrogenic diabetes insipidus. Annu Rev Physiol 2001; 63: 607–630.
Wildin RS, Cogdell DE, Valadez V . AVPR2 variants and V2 vasopressin receptor function in nephrogenic diabetes insipidus. Kidney Int 1998; 54: 1909–1922.
Cooper DN, Krawczak M . The mutational spectrum of single base-pair substitutions causing human genetic disease: patterns and predictions. Hum Genet 1990; 85: 55–74.
Demura M, Takeda Y, Yoneda T, Furukawa K, et al. Two novel types of contiguous gene deletion of the AVPR2 and ARHGAP4 genes in unrelated Japanese kindreds with nephrogenic diabetes insipidus. Hum Mutat 2002; 19: 23–29.
Li J, Daniels GA, Lieber MR . Asymmetric mutation around the recombination break point of immunoglobulin class switch sequences on extrachromosomal substrates. Nucleic Acids Res 1996; 24: 2104–2111.
Schoneberg T, Pasel K, von Baehr V, Schulz A, et al. Compound deletion of the rhoGAP C1 and V2 vasopressin receptor genes in a patient with nephrogenic diabetes insipidus. Hum Mutat 1999; 14: 163–174.
Acknowledgements
We thank the family members for their participation in this study, Fadi Al-Kayal, Mohamed Rajab and Danyah Trabzuni for technical assistance. This study was performed as part of RAC approved Project # 2011056 and during the course of routine molecular diagnostic services, and was not externally funded.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Carroll, P., Al-Mojalli, H., Al-Abbad, A. et al. Novel mutations underlying nephrogenic diabetes insipidus in Arab families. Genet Med 8, 443–447 (2006). https://doi.org/10.1097/01.gim.0000223554.46981.7a
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1097/01.gim.0000223554.46981.7a
Keywords
This article is cited by
-
Novel mutations associated with nephrogenic diabetes insipidus. A clinical-genetic study
European Journal of Pediatrics (2015)
-
Pediatric renal diseases in the Kingdom of Saudi Arabia
World Journal of Pediatrics (2012)
-
A case of aquaporin 2 R85X mutation in a boy with congenital nephrogenic diabetes insipidus
Pediatric Nephrology (2008)