Abstract
Purpose: Polymorphisms in cytokine genes can determine their level of expression and affect the phenotypic expression of immunomediated diseases, such as celiac disease (CD). We thus evaluated cytokine polymorphism prevalence in CD patients and population controls, and assessed the correlation between specific polymorphisms and celiacs' clinical characteristics.
Methods: 155 CD patients and 202 population controls were enrolled in the study. Cytokine polymorphisms (TNFα −308 and −238, IL-1β −511 and +3954, IL-1b RN + 2018, IL-6 −174, IL-10 −1082, −819 and −592, TGFβ1 + 29 and +74, IFN-γ + 874) and HLA class II alleles were determined by sequence-specific primer polymerase chain reaction or restriction fragment length polymorphism analysis. Duodenal histology was classified using Marsh's criteri a. Variables significantly associated with CD patients' characteristics were identified by multivariate logistic regression analysis.
Results: A significantly higher frequency of −308 TNFα polymorphism was observed in CD patients compared to the entire population control group, although no difference was detected when population controls were stratified according to HLA class II. Among celiacs, early onset of the disease (≤2 year old) and the presence of a severe intestinal lesion (Marsh 3C) were significantly associated with the −1082 A/A IL-10 genotype which corresponds to a low producer phenotype (OR 3.28, 95% CI 1.49–7.19 and OR 2.60, 95% CI 1.04–6.49, respectively).
Conclusion: The association between IL-10 genotypes and both histological severity at diagnosis and age of onset could be related to an alteration in cytokine balance, and supports the idea that the various clinical manifestations of the disease could be determined by a different genetic background.
Similar content being viewed by others
Main
Celiac disease (CD) is a common, world-wide disorder characterized by less or more severe atrophy of small bowel villi, crypt hyperplasia and lymphocyte infiltration in lamina propria and epithelium. The damage observed in the intestinal mucosa is the result of the interplay between several genes and gluten, the environmental factor which triggers the immune response in celiac subjects.1
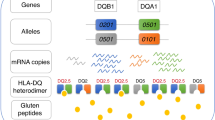
Celiac disease, in 80–90% of the cases, is associated with a specific HLA class II heterodimer, generated by the products of the DQA1*0501 and DQB1*02 alleles (DQ2), whereas the DQA1*03/DQB1*0302 heterodimer (DQ8) is detected in the major part of DQ2-negative cases.2 However, the association with DQ alleles cannot explain the wide range of clinical pictures observed in celiac disease, such as the different age of onset, the variable severity of histological damage or the presence of associated disorders. In fact, it has been calculated that the HLA region contributes for 40% to the family risk of developing celiac disease.3 Several genome wide linkage studies have been performed in order to identify other genomic regions containing disease predisposing variants.4 Among the loci identified, two of them (5q31-33 and 19p13.1) have also been associated to another chronic inflammatory intestinal disease such as Crohn's disease.5 Moreover, a recent paper by Monsuur at al. which analyzed the 19p13.1 region reported a significant association to an intronic variant of the gene MYOSIN IXB, involved in actin remodeling of epithelial enterocytes.6 On the other hand, significant linkage has also been detected for the 2q23-32 region,4 which contains the CD28, CTLA4 and ICOS genes, that regulate the T-cell activation triggered by antigen exposure on HLA class II molecules by antigen presenting cells.7
Involvement of immune system and T-cell activation in the development of the intestinal damage observed in celiac disease has been ascertained by several evidences.8–10 T-cell activation mainly generates a Th1 response characterized by increased production of IL-2, interferon-γ (IFNγ), IL-6 and tumor necrosis factor-α (TNF-α)11,12, which, in turns, triggers cell-mediated immunity and thus intestinal mucosal damage. This response can be counteracted by the production of anti-inflammatory cytokines such as IL-10, which inhibits monocytes/macrophages activation and IFNγ production.13
The production of cytokines can be modulated both by the stimuli present in the local environment as well as by genetic factors.14–18 In fact, the presence of polymorphisms within the coding or noncoding sequences of cytokine genes can alter the efficiency of transcription of these genes and thus the production of cytokines, as demonstrated by studies both in vitro and in vivo.15–18 These polymorphisms could thus play a role in determining the phenotypic expression of CD, shifting the balance toward an increased pro-inflammatory cytokine expression. Up to now, studies performed on CD patients have mainly focused on TNFα promoter polymorphisms, with conflicting results regarding its association to the disease independently from the DQ2 haplotype.19–25 Moreover, very few studies have subdivided celiac patients according to their characteristics and, even in this case, only HLA class II alleles or antibody production have been considered.22–24,26
The aim of this study was to compare the prevalence of polymorphisms of some pro- and anti-inflammatory cytokines genes in a large series of Italian celiac patients and population controls, and to analyze their possible relationship with different clinical parameters.
MATERIALS AND METHODS
Subjects
From April, 2004 to June, 2004 155 consecutive celiac patients (48 males and 107 females) who attended the Gastroenterology Unit at the Department of Medical Sciences, IRCCS – Ospedale Maggiore, Milan, Italy for their yearly follow-up were enrolled in this study. Diagnosis had been established in this Department or in the Pediatric Clinic of Milan University when the patients were ≤2 and >2 year old in 51 and 104 cases, respectively; for patients in the latter subgroup, the mean age at diagnosis was 30.6 ± 17.6 years. All patients had been histologically diagnosed: the adult group underwent duodenal biopsy at diagnosis and 18-24 months after gluten-free diet, whereas the pediatric group was diagnosed according to ESPGHAN criteria, which required the finding of a characteristic small intestine mucosal abnormality at histological examination, clinical remission on gluten-free diet and relapse after gluten challenge.27 All the patients had at least one positive serologic marker of CD (antigliadin, antiendomysial, and antitissue transglutaminase antibodies) at the moment of diagnosis. None of the patients had IgA deficiency. All patients responded to the gluten-free diet, and the histological examination performed on the biopsies obtained during the follow-up endoscopy revealed a Marsh grade 0 or I. HLA typing was performed as part of the diagnostic routine; the DQ2 heterodimer was present in 139 celiac patients (88.5%), whereas other seven carried the DQ8 heterodimer (4.5%). The population control group included 202 adult subjects (98 males and 104 females, mean age years 27.3 ± 18.9), randomly selected from a large study performed on the population from the same geographical area for health surveillance purposes. None of these subjects had gastrointestinal symptoms or associated immune diseases. Forty-three population controls carried the DQ2 heterodimer (21.3%) and five the DQ8 heterodimer (2.5%). All the subjects (patients and population controls) included in the study were of Caucasian origin. All patients gave their informed consent to participate in the study, which was authorized by the local Ethical Committee (IRCCS Ospedale Maggiore, Milan, Italy).
Criteria for CD diagnosis
Serum immunoglobulins were determined by standard techniques in order to exclude IgA deficiency. IgA antigliadin antibodies (AGA) and antitissue transglutaminase antibodies were analyzed with a commercial ELISA kit (OR-GenTec Diagnostika GmbH, Germany and Eu-tTG IgA, Eurospital, Trieste, Italy, respectively), and antiendomysial IgA antibodies (EMA) by indirect immunofluorescence on monkey esophagus (Biognost Endomysiale AK, Bios GmbH Labordiagnostik, Germany). Serological positivity led to intestinal biopsies obtained by Crosby capsule in infants or by endoscopy in older children and adults. Biopsies were processed and stained with hematoxylin-eosine in accordance to standard techniques. The histological description followed Marsh classification, modified by Oberhuber et al.28 Only grade 3 alterations [villous atrophy moderate (A), subtotal (B) or total (C), crypt hyperplasia and increased lymphocytes in lamina propria and epithelium] allowed to diagnose celiac disease. Classification into Marsh' subtypes was determined by the histological damage observed in at least three out of the four biopsies obtained from each patients. Response to gluten-free diet was observed in all patients.
DNA isolation
DNA was extracted from peripheral blood lymphocytes using the Wizard DNA purification kit (Promega, Milan, Italy) according to the manufacturer's instruction.
Cytokine genotyping
The presence of polymorphisms in position −308 (G/A) in the TNFα promoter, −1082 (G/A), −819 (C/T) and −592 (C/A) in the IL-10 promoter and −174 (G/C) in the IL-6 one, as well as in codon 10 (+29, T/C) and codon 25 (+74, G/C) in the TGFβ1 gene and in position +874 (A/T) in intron 1 of the IFN-γ gene were analyzed using the sequence specific primers (SSP)-PCR Cytokine Genotyping kit (One Lamda Inc., Canoga Park, CA) according to the manufacturer's instruction. Polymorphism in position −238 (G/A) in the TNFα promoter was assessed using the primers reported by Fargion et al.;29 the obtained PCR products were then digested with MspI and analyzed on a 2.5% agarose gel. The DNA fragments containing the polymorphisms −511 (C/T) and +3954 (C/T) in the IL-1β gene were amplified as previously described by Nemetz et al.30 and the PCR products digested with AvaI and TaqI, respectively. The IL-1b RN polymorphism in position +2018 (T/C) was assessed as previously described by Clay et al.31
HLA typing
High resolution class II genotyping for the DRB1, DQA1 and DQB1 loci was performed by SSP-PCR using commercial kits according to manufacturer's instructions (Olerup, Uppsala, Sweden and Dynal, Oslo, Norway).
Statistical analysis
Allele and genotype frequencies were compared between cases and controls by chi-square analysis followed, in case of a significant result, by multiple comparisons. Deviations from Hardy-Weinberg equilibrium were tested with an exact test.32 Polymorphisms were analyzed both as nucleotide variations at a single locus or as combined genotypes. The linkage disequilibrium (D) between two loci is expressed as the percentage of the observed disequilibrium with respect to the maximum value (positive or negative as it corresponds).33 Haplotype frequencies were calculated using PHASE software,34 which provides a haplotype reconstruction from population genetic data. Adjusted odds ratios (OR) with 95% confidence intervals (CI) were computed by multivariate logistic regression according to a backward procedure using the SAS statistical software (6.12 version SAS Institute Inc., SAS/STAT User's Guide, Version 6, Fourth Edition, Volume 2, Cary, NC, 1989).
With 155 cases and 202 population controls this study has a power of 0.80 (at least) of demonstrating a 0.15 difference in the prevalence of a characteristic, such as polymorphisms, of cases against population controls or, equivalently, odds ratio ranging from 1.86 to 4.75 at a χ2 test with a significance level of 0.05.
RESULTS
Cytokine polymorphisms in celiac patients and population controls
No significant difference was observed in the genotypic (Table 1 and Table 2) or allelic frequencies of the different cytokine polymorphisms in celiac patients and population controls, apart from the G to A substitution in the homozygous state in position −308 in the TNFα promoter, which was significantly more frequent in celiac patients. This difference was evident even when the −308A allelic frequency was considered (0.274 vs. 0.114 in CD patients and population controls, P < 0.001). However, a significant linkage disequilibrium was detected between the A allele in position −308 in the TNFα promoter and the DQB1*0201, being 82% and 63% in celiac patients and population controls, respectively (P < 0.01 and <0.05). In fact, when the −308A allelic frequency was compared in celiac patients and population controls bearing the DQ2 haplotype, no significant difference could be detected (0.397 vs. 0.352 in celiacs and population controls respectively). Due to the small sample size of DQ8 positive patients and population controls (7 and 5 subjects, respectively), we could not assess whether or not the −308A polymorphism was associated with the presence of the DQA1*03/DQB1*0302 alleles. All the examined alleles, both in celiac patients as well as in population controls, were in Hardy-Weinberg equilibrium, with negligible chi-square values. In all subjects, a 100% linkage disequilibrium was detected between the IL-10 −819 and −592 alleles (P < 0.01).
Association between IL-10 −1082 polymorphism and age of onset of celiac disease
To evaluate the possible association between cytokine polymorphisms and the clinical characteristics of celiac patients, univariate analysis was performed subdividing the patients according to the presence of a positive family history for celiac disease (at least one first degree relative affected), association with other autoimmune diseases, age at onset (≤ or >2 year old) and severity of the intestinal damage as assessed by histology at diagnosis. The 2-year-old cutoff was chosen in order to obtain two classes: one with the maximal expected rate of classic CD (diarrhea, weight loss and growth retardation) and the other with patients with different clinical pictures of CD at presentation (anemia, dyspepsia, hypertransaminasemia).
No difference in the frequency of all polymorphisms, neither considered as genotype nor allele frequency was observed in the subgroups of celiac patients affected by associated autoimmune diseases (N = 41) and in those with familiarity (N = 27), (data not shown).
When celiac patients were subdivided according to the age of onset, a significant difference (P = 0.037) was observed in the frequency of the IL-6 −174C/C genotype. In fact, this genotype was less represented in the 51 (21 males and 30 females) celiac patients ≤2 year old than in the 104 (27 males and 77 females) celiac patients >2 year old (3.9% vs. 17.3% respectively). On the contrary, the frequency of IL-10 −1082 A/A genotype in celiac patients ≤2 year old was significantly higher than that observed in patients with a later onset of the disease (51% vs. 19.2% respectively, P < 0.001). Two other variables were significantly associated with a younger age at diagnosis, namely the male sex (41.2% vs. 25.9% in celiacs ≤ or >2 year old, P = 0.048) and the increased severity of the intestinal damage (Marsh 3C 88.2% vs. 51% in celiacs ≤ or >2 year old, P < 0.001). At multivariate logistic regression analysis, an increased odds ratio was associated with the presence of the IL-10 −1082 A/A genotype, male sex and Marsh 3C lesion, as shown in Table 3, whereas no significant association could be confirmed with the IL-6 −174 C/C genotype.
To assess whether the presence of a single A allele in position −1082 in the IL-10 promoter could be associated with an early onset of the disease, we performed an analysis comparing the A/A, G/A and G/G genotype. Only the A/A genotype showed a significantly increased odds ratio of an early development of the disease compared both to the G/A genotype (OR 3.1545, 95% CI 1.3986–7.2993) and the G/G genotype (OR 3.788, 95% CI 1.145–14.0845), whereas the G/A genotype did not carry an increased risk as compared to the G/G one (OR 1.200, 95% CI 0.379–4.289).
Association between IL-10 −1082 polymorphism and severity of histological damage
When celiac patients were subdivided according to the severity of histological intestinal damage (Marsh <3C N. = 60, 25 males and 35 females; Marsh 3C N = 95, 23 males and 72 females), a significantly higher frequency of the IL-10 −1082 A/A genotype was observed in celiacs with a more severe intestinal lesion (14 vs. 38.8% in Marsh 3A/B and Marsh 3C respectively, P = 0.002). Two other variables were significantly associated with the presence of a Marsh 3C lesion, namely the male sex (24.2% vs. 41.6% in Marsh 3A/B and Marsh 3C respectively, P = 0.037) and the younger age (10.5 vs. 45.9%, P < 0.001). At multivariate analysis, only the presence of the IL-10 −1082 A/A genotype and age ≤2 years old at diagnosis showed significant odds ratios, as shown in Table 4.
Even in this case, multivariate analysis was also performed comparing the A/A, G/A and G/G genotype. The A/A genotype carried a significantly increased risk only as compared to the G/G genotype (OR 3.840, 95% CI 1.188–13.096), whereas the G/A genotype did not (OR 1.657, 95% CI 0.618–4.531).
DISCUSSION
It is now well-known that celiac disease is a disorder due to a partially-explained immunomediated mechanism caused by environmental and genetic factors.1–4 Celiac disease presents with different clinical characteristics ranging from absence of symptoms to a life-threatening condition, different age of onset and variable degree of severity in the intestinal mucosa damage.1 These different clinical manifestations could be explained by the presence of additional modifier genes which may influence the disease phenotype. Candidate genes may include those coding for cytokines, which have been demonstrated to play an important role in the immune response of CD patients.11–13 Several polymorphisms have been detected both in the coding and non-coding regions of these genes, and in vitro and in vivo data showed a correlation between specific polymorphisms and the levels of the produced proteins or the phenotype of various immunomediated diseases.15–18,29,30
In the present study, no significant difference in frequencies of the analyzed polymorphisms could be detected between the CD patients and population controls, apart from a significantly higher frequency of the A allele in position −308 in the TNFα promoter. Comparable frequencies in the polymorphisms in celiacs and population controls have been found also by Wolley et al.,25 who recently evaluated a series of Finnish CD patients using a family-based approach, thus supporting the idea that these polymorphisms per se do not provide an increased susceptibility to CD. The frequency of the −308A allele observed in our series is similar to that observed in other patients from the Mediterranean area,21,22 but lower than that reported in Irish and Swedish series.19,23 However, in our population, no significant difference could be observed when patients and population controls were stratified according to the presence of DQ2, similarly to what reported by Polvi et al.20 and Wolley et al.25
To assess whether cytokine polymorphisms could affect the clinical manifestations of celiac disease we performed a multivariate analysis, considering also the possible combined genotypes. Only the presence of the −1082 polymorphism in the IL-10 promoter resulted significantly associated both with the age of onset and a more severe mucosal lesion. The presence of the G to A substitution at position −1082 in the IL-10 promoter reduces the level of transcription of the IL-10 gene in vitro,15 and in vivo studies demonstrated that the presence of the −1082A allele is associated with reduced quantities of IL-10 both at the mRNA and protein level.16,18,24 IL-10 is a potent anti-inflammatory cytokine which may act at various levels to counteract the effects of pro-inflammatory cytokines such as IFN-γ, IL-2, TNF-α, and IL-4 and to inhibit the antigen-presentation capacity of macrophages and dendritic cells.35 Interestingly, IL-10 polymorphisms with a low production phenotype have been associated with a more severe form of juvenile rheumatoid arthritis,15 disease severity in multiple sclerosis,36 and rate of progression in primary glomerulonephritis.37 In all cases, no differences in polymorphism prevalence were observed between the patient and control populations, again suggesting that these polymorphisms do not represent the causative gene but contribute to modifing the phenotype.
As regards celiac disease, IL-10 polymorphisms have been investigated in different studies.23–26 Two of them did not detect any significant difference in the frequency of these polymorphisms between celiacs and controls,25,26 whereas one observed a significant lower prevalence of low TNFα/high IL-10 producers in celiacs with IgA deficiency versus celiacs with normal IgA levels and controls.24 Interestingly, Hahn-Zoric et al.23 inversely correlated the presence of the low TNFα/high IL-10 producer genotype with the serum levels of antitissue transglutaminase antibodies. Although the role of anti-TG antibodies in the pathogenesis of CD and in the development of the intestinal lesions is still unclear, the higher levels of antitissue transglutaminase antibodies observed in the low IL-10 producers could be attributed to the presence of a more severe intestinal lesion.38,39 The pivotal role of IL-10 in the development of the celiac lesion is also supported by a recent paper by Salvati et al.,13 which demonstrated that IL-10 was able to inhibit the cytokine production triggered by gliadin in duodenal biopsies of CD patients. Lastly, it must be noted that in our present study the A allele in position −1082 in the IL-10 promoter was significantly associated with an earlier diagnosis, possibly suggesting the existence of genetic differences between celiac patients diagnosed in childhood or adulthood. This hypothesis is supported by the recent report by Ashwood et al.,40 who detected a decreased production of IL-10 by intraepithelial and intralamina propria lymphocytes in duodenal biopsies of CD patients diagnosed in pediatric age. This unbalance in the cytokine production toward a pro-inflammatory response could thus cause an earlier development of the disease.
In conclusion, the present study evaluated the frequency of polymorphisms in several cytokine genes in a large series of Italian celiac patients and population controls. For the first time we found a significant association between IL-10 genotypes and both histological severity at diagnosis and age of onset, thus supporting the hypothesis that a different genetic background affects the clinical manifestations of the disease.
References
American Gastroenterological Association medical position statement: celiac sprue. Gastroenterology 2001; 120: 1522–1525.
Sollid LM . Coeliac disease: dissecting a complex inflammatory disorder. Nat. Rev. Immunol 2002; 2: 647–655.
Petronzelli F, Bonamico M, Ferrante P, Grillo R, et al. Genetic contribution of the HLA region to the familial clustering of coeliac disease. Ann Hum Genet 1997; 61: 307–317.
van Heel DA, Hunt K, Greco L, Wijmenga C . Genetics in celiac disease. Best Pract Res Clin Gastroenterol 2005; 19: 323–339.
Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, et al. Genomewide search in anadian families with inflammatory bowel disease reveals two novel susceptibility loci. Am J Hum Genet 2000; 66: 1863–1870.
Monsuur AJ, de Bakker PIW, Alizadeh BZ, Zhernakova A, et al. Myosin IZB variant increases the rish of celiac disease and points toward a primary intestinal barrier defect. Nat Genet 2005; 37: 1341–1344.
Rudd CE, Schneider H . Unifying concepts in CD28, ICOS and CTLA4 co-receptor signalling. Nat rev Immunol 2003; 3: 544–556.
Marsh MN . The morphology and immunopathology of the jejunal lesion in gluten sensitivity. Eur J Gastroenterol Hepatol 2001; 3: 163–168.
Buri C, Burri P, Bahler P, Straumann A, et al. Cytotoxic T-cells are preferentially activated in the duodenal epithelium from patients with florid coeliac disease. J Pathol 2005; 206: 178–185.
Maiuri L, Ciacci C, Ricciardelli I, Vacca L, et al. Association between innate response to gliadin and activation of pathogenic T-cells in coeliac disease. Lancet 2003; 362: 30–37.
Przemioslo RT, Kontakou M, Nobili V, Ciclitira PJ . Raised pro-inflammatory cytokines interleukin 6 and tumor necrosis factor α in coeliac disease mucosa detected by immunohistochemistry. Gut 1994; 35: 1037–1041.
Nilsen EM, Jahnsen FL, Lundin KE, Johansen FE, et al. Gluten induces an intestinal cytokine response strongly dominated by interferon gamma in patients with celiac disease. Gastroenterology 1998; 115: 551–563.
Salvati VM, Mazzarella G, Gianfrani C, Levings MK, et al. Recombinant human interleukin 10 suppresses gliadin dependent T-cell activation in ex vivo cultured coeliac intestinal mucosa. Gut 2005; 54: 46–53.
Hawiger J . Innate immunity and inflammation: a transcriptional paradigm. Immunol Res 2001; 23: 99–109.
Crawley E, Kay R, Sillibourne J, Patel P, et al. Polymorphic haplotypes of the interleukin-10 5′ flanking region determine variable interleukin-10 transcription and are associated with particular phenotypes of juvenile rheumatoid arthritis. Arthritis Rheum 1999; 42: 1101–1108.
Koss K, Satsangi J, Fanning GC, Welsh KI, et al. Cytokine (TNF alpha, LT alpha and IL-10) polymorphisms in inflammatory bowel diseases and normal controls: differential effects on production and allele frequencies. Genes Immun 2000; 1: 185–190.
Hulkkonen J, Pertovaara M, Antonen J, Pasternack A, et al. Elevated interleukin-6 plasma levels are regulated by the promoter region polymorphism of the IL6 gene in primary Sjogren's syndrome and correlate with the clinical manifestations of the disease. Rheumatology 2001; 40: 656–661.
Rad R, Dossumbekova A, Neu B, Lang R, et al. Cytokine gene polymorphisms influence mucosal cytokine expression, gastric inflammation, and host specific colonisation during Helicobacter pylori infection. Gut 2004; 53: 1082–1089.
McManus R, Wilson AG, Mansfield J, Weir DG, et al. TNF2, a polymorphism of the tumour necrosis-alpha gene promoter, is a component of the celiac disease major histocompatibility complex haplotype. Eur J Immunol 1996; 26: 2113–2118.
Polvi A, Maki M, Collin P, Partanen J . TNF microsatellite alleles a2 and b3 are not primarily associated with celiac disease in the Finnish population. Tissue Antigens 1998; 51: 553–555.
de la Concha EG, Fernandez-Arquero M, Vigil P, Rubio A, et al. Celiac disease and TNF promoter polymorphisms. Hum Immunol 2000; 61: 513–517.
Garrote JA, Arranz E, Telleria JJ, Castro J, et al. TNF alpha and LT alpha gene polymorphisms as additional markers of celiac disease susceptibility in a DQ2-positive population. Immunogenetics 2002; 54: 551–555.
Hahn-Zoric M, Hytonen AM, Hanson LA, Nilsson LA, et al. Association of −1087 IL10 and −308 TNFΑ gene polymorphisms with serological markers of coeliac disease. J Clin Immunol 2003; 23: 291–296.
Cataldo F, Lio D, Marino V, Scola L, et al. Groups of the SIGEP and “Club del Tenue”. Cytokine genotyping (TNF and IL-10) in patients with celiac disease and selective IgA deficiency. Am J Gastroenterol 2003; 98: 850–856.
Wolley N, Mustalahti K, Maki M, Partanen J . Cytokine gene polymorphisms and genetic association with coeliac disease in the Finnish population. Scand J Immunol 2005; 61: 51–56.
Garrote J, Arranz E, Gomez-Gonzalez E, Leon A, et al. IL6, IL10 and TGFB1 gene polymorphisms in celiac disease: differences between DQ2 positive and negative patients. Allergol Immunopathol 2005; 33: 245–249.
Revised criteria for diagnosis of coeliac disease. Report of Working Group of European Society of Paediatric Gastroenterology and Nutrition. Arch Dis Child 1990; 65: 909–911.
Oberhuber G, Granditsch G, Vogelsang H . The histopathology of coeliac disease: time for a standardized report scheme for pathologists. Eur J Gastroenterol Hepatol 1999; 11: 1185–1194.
Fargion S, Valenti L, Dongiovanni P, Scaccabarozzi A, et al. Tumor necrosis factor alpha promoter polymorphisms influence the phenotypic expression of hereditary hemochromatosis. Blood 2001; 97: 3707–3712.
Nemetz A, Toth M, Garcia-Gonzalez MA, Zagoni T, et al. Allelic variation at the interleukin 1 beta gene is associated with decreased bone mass in patients with inflammatory bowel diseases. Gut 2001; 49: 644–649.
Clay FE, Tarlow JK, Cork MJ, Cox A, et al. Novel interleukin-1 receptor antagonist exon polymorphisms and their use in allele-specific mRNA assessment. Hum Genet 1996; 97: 723–726.
Elston RC, Forthofer R . Testing for Hardy-Weinberg equilibrium in small samples. Biometrics 1997; 33: 536–542.
Hartl DL, Clark AG . Principles of population genetics. Second edition. Sunderland, MA: Sinauer Associates Inc., 1989; 1–60.
Stephens M, Smith N, Donnelly P . A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 2001; 68: 978–989.
Pestka S, Krause CD, Sarkar D, Walter MR, et al. Interleukin 10 and related cytokines and receptors. Annu Rev Immunol 2004; 22: 929–979.
Luomala M, Lehtimaki T, Huhtala H, Ukkonen M, et al. Promoter polymorphism of IL-10 and severity of multiple sclerosis. Acta Neurol Scand 2003; 108: 396–400.
Bantis C, Heering PJ, Aker S, Klein-Vehne N, et al. Association of interleukin-10 gene G-1082A polymorphism with the progression of primary glomerulonephritis. Kidney Int 2004; 66: 288–294.
Freitag T, Schulze-Koops H, Niedobitek G, Melino G, et al. The role of the immune response against tissue transglutaminase in the pathogenesis of coeliac disease. Autoimmunity Reviews 2004; 3: 13–20.
Korponay-Szabò IR, Halttunen T, Szalai Z, Laurila K, et al. In vivo targeting of intestinal and extraintestinal transglutaminase 2 by coeliac autoantibodies. Gut 2004; 53: 641–648.
Ashwood P, Anthony A, Torrente F, Wakefield AJ . Spontaneous mucosal lymphocyte cytokine profiles in children with autism and gastrointestinal symptoms: mucosal immune activation and reduced counter regulatory interleukin-10. J Clin Immunol 2004; 24: 664–673.
Acknowledgements
This work was supported by a research grant from IRCCS-Ospedale Maggiore, Milan, Italy and by a grant from the Italian Ministry of Health (ex art. 12) and Regione Lombardia.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Barisani, D., Ceroni, S., Meneveri, R. et al. IL-10 polymorphisms are associated with early-onset celiac disease and severe mucosal damage in patients of Caucasian origin. Genet Med 8, 169–174 (2006). https://doi.org/10.1097/01.gim.0000204464.87540.39
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1097/01.gim.0000204464.87540.39