Abstract
Purpose
Implementation science offers methods to evaluate the translation of genomic medicine research into practice. The extent to which the National Institutes of Health (NIH) human genomics grant portfolio includes implementation science is unknown. This brief report’s objective is to describe recently funded implementation science studies in genomic medicine in the NIH grant portfolio, and identify remaining gaps.
Methods
We identified investigator-initiated NIH research grants on implementation science in genomic medicine (funding initiated 2012–2016). A codebook was adapted from the literature, three authors coded grants, and descriptive statistics were calculated for each code.
Results
Forty-two grants fit the inclusion criteria (~1.75% of investigator-initiated genomics grants). The majority of included grants proposed qualitative and/or quantitative methods with cross-sectional study designs, and described clinical settings and primarily white, non-Hispanic study populations. Most grants were in oncology and examined genetic testing for risk assessment. Finally, grants lacked the use of implementation science frameworks, and most examined uptake of genomic medicine and/or assessed patient-centeredness.
Conclusion
We identified large gaps in implementation science studies in genomic medicine in the funded NIH portfolio over the past 5 years. To move the genomics field forward, investigator-initiated research grants should employ rigorous implementation science methods within diverse settings and populations.
Similar content being viewed by others
Introduction
The rate of translation of genomic discoveries to benefit patient and population health has been slow compared with the rate of discovery.1,2 As such, the majority of genomic research falls within the discovery and development phases (T0–T1), and only 2% of research falls within the translational phases (T2–T4).3 Implementation science (IS) is a field of research that examines methods and strategies that aim to improve the translation of research discoveries to practice settings, making IS well suited to speed the rate of translation of genomic discoveries to benefit patient and population health.4
An increasing number of applications in genomics have been included in evidence-based guidelines and are improving patient health.4 For those evidence-based genomics applications, implementation research can improve their translation into clinical and public health practice to improve health. For developing genomic applications, implementation should be considered across the research continuum; by planning for implementation early, the length of time from bench to bedside may be reduced once the evidence base for the application has accrued.
In a recent literature review,5 we examined the extent to which translational genomic medicine research has incorporated IS methods. In the review, we identified several important gaps in the current literature, including a lack of rigorous IS methods (e.g., suboptimal use of IS conceptual frameworks), lack of attention to IS components such as capacity building and sustainability, low reporting of race and ethnicity as well as a lack of diversity in study populations and settings, and finally, the fact that most studies were descriptive and within the field of oncology. It remains unclear to what extent the National Institutes of Health (NIH) portfolio of funded grants will address some of these identified gaps in the literature. As such, the objective of this brief report is to examine the current NIH grant portfolio to (i) ascertain whether recently funded NIH grants are bridging identified gaps from the literature, and (ii) determine what gaps in the NIH portfolio persist.
Materials and methods
NIH extramural grants funded in fiscal years 2012–2016 were identified on 30 September 30 2016 through an internal NIH tool, Query, View, Report (QVR). QVR allows users to search, view, and retrieve detailed information about NIH applications and awards.
Search process
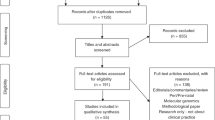
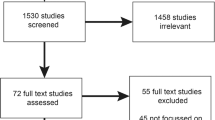
We used QVR to create a search strategy of weighted terms, or “custom fingerprints,” to identify genomic medicine grants that included IS approaches. In our first step, we created three custom fingerprints, designed and performed by three authors (A.E.K., M.C., M.C.R.; Supplementary Table S1 online). These fingerprints were based on modified search terms from grants that were (i) submitted in three Implementing Genomics in Practice (IGNITE)6 funding announcements (n = 729) (A.E.K.); (ii) reviewed by the Dissemination and Implementation Research in Health study group and combined with a separate search using the Research, Condition, and Disease Category term “human genome” (n = 1,557) (M.C.); and (iii) reviewed by the Dissemination and Implementation Research in Health study group with genomics terms, including human genome, genomic medicine, personalized medicine, precision genomic medicine, genetic, and genetic medicine (n = 494) (M.R.). Grants ascertained from the searches were combined, de-duplicated, and reviewed for inclusion (by abstracts and specific aims) to include 508 grants (Table 1). In our second step, we created a custom fingerprint in QVR based on these 508 grants, and we identified an additional, relevant 427 grants. As a final step in the search process, we searched for all grants using the terms “human genome” and “human subjects” in QVR, and we identified an additional 51 relevant grants (total n = 986).
After restricting the set to select awarded, investigator-initiated, research, and career development grants (R01, R03, R21, R33, K01, K07, K23, and K99; n = 154), a review of each application’s research strategies was performed. Upon full review, additional grants were excluded based on inclusion and exclusion criteria7 (Table 1). Our final analytic sample included 42 grants that were coded.
Coding methods
The initial codebook was adapted from a NIH portfolio review of IS funded studies, with additions and modifications made based upon our previous literature analysis of IS studies in the translational genomic medicine literature.5 A subset of grants (n = 9) were triple-coded by all three authors (A.E.K., M.C., M.C.R.). Coding discrepancies were discussed and agreement was reached with additional comments and clarification added to the codebook to establish coding consistency. The remaining grants were divided for individual coding by the three authors. Questions about coding that occurred during this process were addressed and resolved by all three coders through consensus. To assess quality control across the single-coded grants, one author retrospectively coded a random sample of 20% of grants and found 92% agreement in coding. Additional quality control checks and review were performed on data when the single-coded data was merged into the final analytic file.
As a secondary analysis, we separately described cooperative agreements funded during the same period (2012–2016) with an adapted codebook. One author abstracted information about study design (i.e., study setting), genomics (i.e., disease area), and IS (i.e., use of IS frameworks and capacity indicators); a second author checked for consistency of coding; and questions about inclusion were discussed until consensus was reached.
Analysis
Codes were summed, and descriptive statistics (i.e., proportions, means) were calculated.
Results
We identified 42 genomic medicine grants that included elements of IS, representing approximately 1.75% of the investigator-initiated research grants in genomics (Table 2,Supplementary Table S2). Most included grants included T3 research (81%, n = 36), while only 12% included T2 phase research, which represented “pre-implementation” focused evaluation research (i.e., research primarily focused on validity and utility, but in this case, also included some IS component). Only one grant included T4 research.
Grant study design characteristics
Most grants proposed qualitative (n = 35, 83.3%) and/or quantitative methods (n = 40, 95.2%), including cost analyses (n = 4), comparative effectiveness (n = 3), and simulation modeling (n = 1). Furthermore, study designs primarily included cross-sectional designs (n = 25), but also included cohort (n = 9), randomized controlled trials (n = 9), pre/post (n = 7), and case-control (n = 1) designs. Most awarded grants proposed research within the clinical setting (n = 32, 76.2%) rather than public health settings (n = 4, 9.5%), or other settings (e.g., online) (n = 5, 11.9%). Proposed study samples primarily consisted of white (average proportion of whites = 71.3%, median proportion of whites = 75.6%), non-Hispanic (average proportion of Hispanics = 81.2%, median proportion of Hispanics = 88.9%) study populations.
Genomic research focus
Few grants focused on family history collection (11.9%, n = 5). Instead, most described germ-line genetic testing (73.8%, n = 31), with a minority of grants focusing on somatic (9.5%, n = 4) or cell-free DNA testing (7.1%, n = 3). More specifically, awarded grants included research on single-gene tests (21.4%, n = 9), whole-genome sequencing (14.3%, n = 6), whole-exome sequencing (n = 5, 11.9%), or gene panel testing (n = 5, 11.9%). Half of the grants focused on the use of genomic medicine for risk assessment (n = 21), and fewer included research aims related to diagnostic (n = 11, 30%), therapeutic (n = 7, 26.1%), preventive (n = 4, 9.5%), or prognostic (n = 1, 2.4%) testing. Most awarded grants included a focus on cancer screening or treatment (n = 19, 45.2%) as opposed to other disease areas, such as newborn screening (n = 3, 7.1%), prenatal testing (n = 3, 7.1%), or other diseases/disorders (e.g., cardiovascular health, general pharmacogenomics, undiagnosed diseases, autism, Huntington disease, kidney disease, psychosis, hearing loss) (40.6%). Finally, many grants proposed to assess patient (54.8%) and provider (21.4%) attitudes, including assessment of barriers and facilitators to the implementation of genomic medicine (n = 7, 16.7%).
Implementation research focus
Most IS in genomic medicine grants had aims related to implementation (n = 37, 88.1%) rather than dissemination(s) (n = 12, 28.6%) or adoption (n = 2, 4.8%). Nine grants included sustainability indicators, such as costs (n = 5, 11.9%), capacity building (n = 3, 7.1%), or maintenance (n = 1, 2.4%) measures. Only four grants used conceptual models from the IS field, with all using the diffusion of innovations model.8 Half (n = 2) used this model for formative research, one used the model for intervention design, and the other grant used the model for measurement.
While most grants did not explicitly include collaborative processes, two grants included designing for dissemination, five included patient engagement and five included stakeholder engagement, two grants included team science approaches, and one included community-based participatory research. Measured implementation and process outcomes included patient-centeredness (e.g., assessing patient barriers and facilitators) (n = 22, 52.4%), uptake (n = 14, 33.3%), feasibility (n = 11, 26.2%), effectiveness (n = 10, 23.8%), acceptability (n = 6, 14.3%), costs (n = 2 monetary, n = 1 nonmonetary, n = 1 both, 9.5%), fidelity (n = 3, 7.1%), equity (n = 3, 7.1%), and efficiency (n = 3 7.1%). Most studies included an individual unit of analysis (n = 35, 83.3%), while three (7.1%) analyzed at the level of the study site (and unit of analysis was unclear in three grants).
In our secondary analysis, similar gaps were found among funded cooperative agreements (data not shown). For example, among the 39 cooperative agreements funded in IS in genomic medicine between 2012 and 2016, only 5% used IS frameworks, most studied oncology (n = 10), most occurred in the clinical setting (n = 34), and while slightly more U grants included measurement of capacity indicators (primarily by measuring costs), only approximately 30% included these indicators.
Discussion
We found that the currently funded implementation research in genomic medicine includes primarily T3 implementation research in clinical settings that focuses on germ-line testing, risk assessment, and oncology. Like the published literature, only one grant included T4 research, suggesting a remaining gap in moving the field forward through all translational research phases.
The study designs proposed in these grant awards were typically cross-sectional, used an individual level unit of analysis, incorporated quantitative and qualitative methods, and occurred within clinical settings. Furthermore, few studies included simulation, cost, and comparative effectiveness analyses. These characteristics largely reflect those found in the current literature. However, funded grants may partially close certain gaps. For example, the funded grant proposals incorporated qualitative methods into their study designs more than the current literature. The racial/ethnic diversity of populations in funded grants was similar to that reported in the current literature, being primarily white, non-Hispanic; however, unlike the published literature, which often lacked information about the racial/ethnic composition of their study populations, information about the racial composition of study populations was reported for all grants per reporting rules for human subjects research at the NIH. The majority of funded awards proposed the use of clinical settings; however, this proportion was even larger among the funded grants, perhaps because many NIH grants are funded to academic institutions within clinical settings. Like the published literature, most grants include cross-sectional study designs; however, more randomized controlled trials and pre/post studies were found among the funded grants than the current literature, suggesting that the body of research is beginning to shift from descriptive and exploratory studies to interventions within clinical and, to a lesser extent, public health settings.
Findings from this portfolio analysis were similar to the current IS literature in genomic medicine, which has primarily focused on germ-line testing to assess cancer risk. Further, the proportion of funded grants examining risk assessment and/or oncology was even greater than the current research literature, despite there being more variation in the type of genomic technology (i.e., germ line, somatic, cell-free DNA) studied among the funded grants. This suggests a sustained gap in research examining applications of genomic medicine to disease areas outside of oncology and applications beyond risk assessment, such as prevention, prognosis, diagnosis, or therapeutic settings (e.g., pharmacogenomics).
Finally, the majority of the literature did not include sustainability measures or incorporate IS conceptual frameworks; the same was true for funded grants, though rates of including sustainability measures and conceptual frameworks were higher in funded grants (e.g., 7.1% sustainability indicators in the literature versus 20% in the funded grants), perhaps due to the presence of the Dissemination and Implementation Research in Health study section, which specifically reviews and awards IS grant applications on metrics including rigorous IS research methods.9 Finally, like the literature, the most frequently measured IS and process outcomes were patient centeredness (often through the collection of barriers and facilitators) and uptake. Incorporating more rigorous IS methods and measures will allow practitioners and researchers to more effectively translate evidence-based genomic discoveries to the benefit patient care.
While this portfolio analysis presents an overview of the currently funded IS in genomic medicine research, the analysis does have limitations. While our multipronged search was comprehensive, it is possible that we missed funded grants that include IS in translational genomic research. Of note, this review does not include grants whose initial funding began prior to 2012. As such, we did not include grants resulting from Clinical Sequencing Exploratory Research (CSER),10 which seeks to translate genomics into clinical practice. CSER2 will extend the efforts of CSER to promote IS studies in genomic medicine.11 The IGNITE consortium was created to enhance the implementation of genomic medicine by supporting the development of methods for incorporating genomic information into clinical care.6 These consortia are intended to fill gaps in the implementation of genomic medicine research, and their success should be evaluated in future studies. Such cooperative agreements were not included in the primary analysis, as we only examined independent research awards. We did not include these grants in the primary analysis because (i) our objective was to describe grants that investigators are submitting, rather than evaluating the success of funding announcements to award IS grants in genomic medicine, and (ii) the scope of cooperative agreements differs from investigator-initiated grants; thus, a different codebook would need to be developed, and findings may not be directly comparable with investigator-initiated grants. Trends in gaps appeared to traverse the investigator-initiated grants and cooperative agreements. Finally, there are other agencies that have funded and can fund IS work, including the Agency for Healthcare Research and Quality, the Patient-Centered Outcomes Research Institute, and the Centers for Disease Control and Prevention, among others. This portfolio analysis does not provide a snapshot of IS research being funded outside of the NIH; however, funding from these agencies is outside the scope of this work. For these reasons, this analysis may underrepresent the currently funded IS studies in genomic medicine; however, we would anticipate that the research gaps identified in this analysis would be similar.
Overall, this portfolio analysis demonstrates a continued need for research at the intersection of IS and genomic medicine. The NIH-wide Dissemination and Implementation Research in Health funding announcement explicitly mentions genomics, and provides another vehicle for funding research at the intersection between genomic medicine and implementation science.12 Moving forward, research that employs rigorous IS methods and measures and examines diverse genomic technologies will help move the translation of genomic medicine to improve patient care and ultimately population health.
References
Khoury MJ, Gwinn M, Yoon PW, Dowling N, Moore CA, Bradley L. The continuum of translation research in genomic medicine: how can we accelerate the appropriate integration of human genome discoveries into health care and disease prevention? Genet Med 2007;9:665–674.
Manolio TA, Chisholm RL, Ozenberger B et al. Implementing genomic medicine in the clinic: the future is here. Genet Med 2013;15:258–267.
Clyne M, Schully SD, Dotson WD et al. Horizon scanning for translational genomic research beyond bench to bedside. Genet Med 2014;16:535–538.
Chambers DA, Feero WG, Khoury MJ. Convergence of implementation science, precision medicine, and the learning health care system: a new model for biomedical research. JAMA 2016;315:1941–1942.
Roberts MC, Kennedy AE, Chambers DA, Khoury MJ. The current state of implementation science in genomic medicine: opportunities for improvement. Genet Med 2017;19:858–863.
Implementing Genomics in Practice (IGNITE). Division of Genomic Medicine Research Programs. https://www.genome.gov/27554264/. Accessed 26 May 2017.
Neta G, Sanchez MA, Chambers DA et al. Implementation science in cancer prevention and control: a decade of grant funding by the National Cancer Institute and future directions. Implement Sci 2015;10:4.
Rogers EM. Diffusion of Innovations. Free Press: New York, 1995.
Proctor EK, Powell BJ, Baumann AA, Hamilton AM, Santens RL. Writing implementation research grant proposals: ten key ingredients. Implement Sci 2012;7:96.
Clinical Sequencing Exploratory Research (CSER)https://cser-consortium.org/. Accessed 8 June 2017.
Clinical Sequencing Evidence-Generating Research (CSER2)—Clinical Sites (U01)https://grants.nih.gov/grants/guide/rfa-files/RFA-HG-16-010.html. Accessed 8 June 2017.
Dissemination and Implementation Research in HealthDepartment of Health and Human Services. National Institutes of Health, 2016. https://grants.nih.gov/grants/guide/pa-files/PAR-16-238.html. Accessed October 17, 2017.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention or the Department of Health and Human Services. The views and opinions expressed in this article are those of the authors and do not represent the views of the Center for Disease Control and Prevention or the Department of Health and Human Services.
Disclosure At the time of this work, M.C.R. was a Cancer Prevention postdoctoral Fellow at the National Cancer Institute. The other authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Roberts, M.C., Clyne, M., Kennedy , A.E. et al. The current state of funded NIH grants in implementation science in genomic medicine: a portfolio analysis. Genet Med 21, 1218–1223 (2019). https://doi.org/10.1038/gim.2017.180
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2017.180
Keywords
This article is cited by
-
Navigating the field of implementation science towards maturity: challenges and opportunities
Implementation Science (2024)
-
What is the power of a genomic multidisciplinary team approach? A systematic review of implementation and sustainability
European Journal of Human Genetics (2024)
-
Evaluating cancer genetic services in a safety net system: overcoming barriers for a lasting impact beyond the CHARM research project
Journal of Community Genetics (2023)
-
Opportunities, challenges and expectations management for translating biobank research to precision medicine
European Journal of Epidemiology (2020)