Abstract
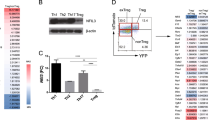
The T-cell immunoglobulin and mucin domain-containing protein 3 (TIM-3) is selectively expressed on terminally differentiated T helper 1 (Th1) cells and acts as a negative regulator that terminates Th1 responses. The dysregulation of TIM-3 expression on T cells is associated with several autoimmune phenotypes and with chronic viral infections; however, the mechanism of this regulation is unclear. In this study, we investigated the effect of DNA methylation on the expression of TIM-3. By analyzing the sequences of TIM-3 promoter regions in human and mouse, we identified a CpG island within the TIM-3 promoter and demonstrated that the promoter activity was controlled by DNA methylation. Furthermore, treatment with 5-aza-2′-deoxycytidine enhanced TIM-3 expression on mouse primary CD4+ T cells under Th0-, Th1- or Th2-polarizing conditions. Finally, pyrosequencing analysis revealed that the methylation level of the TIM-3 promoter gradually decreased after each round of T-cell polarization, and this decrease was inversely correlated with TIM-3 expression. These data suggest that the DNA methylation of the TIM-3 promoter cooperates with lineage-specific transcription factors in the control of Th-cell development. In conclusion, DNA methylation-based regulation of TIM-3 may provide novel insights into understanding the dysregulation of TIM-3 expression under pathogenic conditions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Monney L, Sabatos CA, Gaglia JL, Ryu A, Waldner H, Chernova T et al. Th1-specific cell surface protein TIM-3 regulates macrophage activation and severity of an autoimmune disease. Nature 2002; 415: 536–541.
Zhu C, Anderson AC, Schubart A, Xiong H, Imitola J, Khoury SJ et al. The TIM-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat Immunol 2005; 6: 1245–1252.
Kashio Y, Nakamura K, Abedin MJ, Seki M, Nishi N, Yoshida N et al. Galectin-9 induces apoptosis through the calcium-calpain-caspase-1 pathway. J Immunol 2003; 170: 3631–3636.
Chou FC, Shieh SJ, Sytwu HK . Attenuation of Th1 response through galectin-9 and T-cell Ig mucin 3 interaction inhibits autoimmune diabetes in NOD mice. Eur J Immunol 2009; 39: 2403–2411.
Seki M, Oomizu S, Sakata KM, Sakata A, Arikawa T, Watanabe K et al. Galectin-9 suppresses the generation of Th17, promotes the induction of regulatory T cells, and regulates experimental autoimmune arthritis. Clin Immunol 2008; 127: 78–88.
Niwa H, Satoh T, Matsushima Y, Hosoya K, Saeki K, Niki T et al. Stable form of galectin-9, a TIM-3 ligand, inhibits contact hypersensitivity and psoriatic reactions: a potent therapeutic tool for Th1- and/or Th17-mediated skin inflammation. Clin Immunol 2009; 132: 184–194.
Chou FC, Kuo CC, Wang YL, Lin MH, Linju Yen B, Chang DM et al. Overexpression of galectin-9 in islets prolongs grafts survival via downregulation of Th1 responses. Cell Transplant 2013; 22: 2135–2145.
He W, Fang Z, Wang F, Wu K, Xu Y, Zhou H et al. Galectin-9 significantly prolongs the survival of fully mismatched cardiac allografts in mice. Transplantation 2009; 88: 782–790.
Wang F, He W, Yuan J, Wu K, Zhou H, Zhang W et al. Activation of TIM-3-galectin-9 pathway improves survival of fully allogeneic skin grafts. Transpl Immunol 2008; 19: 12–19.
Koguchi K, Anderson DE, Yang L, O'Connor KC, Kuchroo VK, Hafler DA . Dysregulated T cell expression of TIM3 in multiple sclerosis. J Exp Med 2006; 203: 1413–1418.
Li H, Wu K, Tao K, Chen L, Zheng Q, Lu X et al. TIM-3/galectin-9 signaling pathway mediates T-cell dysfunction and predicts poor prognosis in patients with hepatitis B virus-associated hepatocellular carcinoma. Hepatology 2012; 56: 1342–1351.
Liberal R, Grant CR, Holder BS, Ma Y, Mieli-Vergani G, Vergani D et al. The impaired immune regulation of autoimmune hepatitis is linked to a defective galectin-9/tim-3 pathway. Hepatology 2012; 56: 677–686.
Jones RB, Ndhlovu LC, Barbour JD, Sheth PM, Jha AR, Long BR et al. TIM-3 expression defines a novel population of dysfunctional T cells with highly elevated frequencies in progressive HIV-1 infection. J Exp Med 2008; 205: 2763–2779.
McMahan RH, Golden-Mason L, Nishimura MI, McMahon BJ, Kemper M, Allen TM et al. TIM-3 expression on PD-1+ HCV-specific human CTLs is associated with viral persistence, and its blockade restores hepatocyte-directed in vitro cytotoxicity. J Clin Invest 2010; 120: 4546–4557.
Golden-Mason L, Palmer BE, Kassam N, Townshend-Bulson L, Livingston S, McMahon BJ et al. Negative immune regulator TIM-3 is overexpressed on T cells in hepatitis C virus infection and its blockade rescues dysfunctional CD4+ and CD8+ T cells. J Virol 2009; 83: 9122–9130.
Zhang J, Daley D, Akhabir L, Stefanowicz D, Chan-Yeung M, Becker AB et al. Lack of association of TIM3 polymorphisms and allergic phenotypes. BMC Med Genet 2009; 10: 62.
Chae SC, Park YR, Lee YC, Lee JH, Chung HT . The association of TIM-3 gene polymorphism with atopic disease in Korean population. Hum Immunol 2004; 65: 1427–1431.
Chae SC, Park YR, Shim SC, Yoon KS, Chung HT . The polymorphisms of Th1 cell surface gene TIM-3 are associated in a Korean population with rheumatoid arthritis. Immunol Lett 2004; 95: 91–95.
Gao PS, Mathias RA, Plunkett B, Togias A, Barnes KC, Beaty TH et al. Genetic variants of the T-cell immunoglobulin mucin 1 but not the T-cell immunoglobulin mucin 3 gene are associated with asthma in an African American population. J Allergy Clin Immunol 2005; 115: 982–988.
Zhang CC, Wu JM, Cui TP, Wang P, Pan SX . [Study on relationship between polymorphism sites of TIM-3 and allergic asthma in a population of adult Hans from Hubei province of China]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2006; 23: 74–77.
Bruck P, Ramos-Lopez E, Bartsch W, Bohme A, Badenhoop K . TIM-3 polymorphisms in type 1 diabetes families. J Hum Genet 2008; 53: 559–564.
Anderson AC, Lord GM, Dardalhon V, Lee DH, Sabatos-Peyton CA, Glimcher LH et al. T-bet, a Th1 transcription factor regulates the expression of TIM-3. Eur J Immunol 2010; 40: 859–866.
Wilson CB, Rowell E, Sekimata M . Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol 2009; 9: 91–105.
Baumjohann D, Ansel KM . MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nat Rev Immunol 2013; 13: 666–678.
Bird JJ, Brown DR, Mullen AC, Moskowitz NH, Mahowald MA, Sider JR et al. Helper T cell differentiation is controlled by the cell cycle. Immunity 1998; 9: 229–237.
Young HA, Ghosh P, Ye J, Lederer J, Lichtman A, Gerard JR et al. Differentiation of the T helper phenotypes by analysis of the methylation state of the IFN-gamma gene. J Immunol 1994; 153: 3603–3610.
Valapour M, Guo J, Schroeder JT, Keen J, Cianferoni A, Casolaro V et al. Histone deacetylation inhibits IL4 gene expression in T cells. J Allergy Clin Immunol 2002; 109: 238–245.
Klug M, Rehli M . Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics 2006; 1: 127–130.
Owaki T, Asakawa M, Morishima N, Hata K, Fukai F, Matsui M et al. A role for IL-27 in early regulation of Th1 differentiation. J Immunol 2005; 175: 2191–2200.
Kanno Y, Vahedi G, Hirahara K, Singleton K, O'Shea JJ . Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annu Rev Immunol 2012; 30: 707–731.
Sanchez-Fueyo A, Tian J, Picarella D, Domenig C, Zheng XX, Sabatos CA et al. TIM-3 inhibits T helper type 1-mediated auto- and alloimmune responses and promotes immunological tolerance. Nat Immunol 2003; 4: 1093–1101.
Sabatos CA, Chakravarti S, Cha E, Schubart A, Sanchez-Fueyo A, Zheng XX et al. Interaction of TIM-3 and TIM-3 ligand regulates T helper type 1 responses and induction of peripheral tolerance. Nat Immunol 2003; 4: 1102–1110.
Vali B, Jones RB, Sakhdari A, Sheth PM, Clayton K, Yue FY et al. HCV-specific T cells in HCV/HIV co-infection show elevated frequencies of dual TIM-3/PD-1 expression that correlate with liver disease progression. Eur J Immunol 2010; 40: 2493–2505.
Klibi J, Niki T, Riedel A, Pioche-Durieu C, Souquere S, Rubinstein E et al. Blood diffusion and Th1-suppressive effects of galectin-9-containing exosomes released by Epstein-Barr virus-infected nasopharyngeal carcinoma cells. Blood 2009; 113: 1957–1966.
Anderson AC, Anderson DE, Bregoli L, Hastings WD, Kassam N, Lei C et al. Promotion of tissue inflammation by the immune receptor TIM-3 expressed on innate immune cells. Science 2007; 318: 1141–1143.
Chen Y, Langrish CL, McKenzie B, Joyce-Shaikh B, Stumhofer JS, McClanahan T et al. Anti-IL-23 therapy inhibits multiple inflammatory pathways and ameliorates autoimmune encephalomyelitis. J Clin Invest 2006; 116: 1317–1326.
Penix LA, Sweetser MT, Weaver WM, Hoeffler JP, Kerppola TK, Wilson CB . The proximal regulatory element of the interferon-gamma promoter mediates selective expression in T cells. J Biol Chem 1996; 271: 31964–31972.
Bruniquel D, Schwartz RH . Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol 2003; 4: 235–240.
Fitzpatrick DR, Shirley KM, McDonald LE, Bielefeldt-Ohmann H, Kay GF, Kelso A . Distinct methylation of the interferon gamma (IFN-gamma) and interleukin 3 (IL-3) genes in newly activated primary CD8+ T lymphocytes: regional IFN-gamma promoter demethylation and mRNA expression are heritable in CD44(high)CD8+ T cells. J Exp Med 1998; 188: 103–117.
Makar KW, Wilson CB . DNA methylation is a nonredundant repressor of the Th2 effector program. J Immunol 2004; 173: 4402–4406.
Sellars M, Huh JR, Day K, Issuree PD, Galan C, Gobeil S et al. Regulation of DNA methylation dictates Cd4 expression during the development of helper and cytotoxic T cell lineages. Nat Immunol 2015; 16: 746–754.
Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 2001; 15: 763–774.
Makar KW, Perez-Melgosa M, Shnyreva M, Weaver WM, Fitzpatrick DR, Wilson CB . Active recruitment of DNA methyltransferases regulates interleukin 4 in thymocytes and T cells. Nat Immunol 2003; 4: 1183–1190.
Gamper CJ, Agoston AT, Nelson WG, Powell JD . Identification of DNA methyltransferase 3a as a T cell receptor-induced regulator of Th1 and Th2 differentiation. J Immunol 2009; 183: 2267–2276.
Baron U, Floess S, Wieczorek G, Baumann K, Grutzkau A, Dong J et al. DNA demethylation in the human FOXP3 locus discriminates regulatory T cells from activated FOXP3(+) conventional T cells. Eur J Immunol 2007; 37: 2378–2389.
Ohkura N, Hamaguchi M, Morikawa H, Sugimura K, Tanaka A, Ito Y et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity 2012; 37: 785–799.
Youngblood B, Oestreich KJ, Ha SJ, Duraiswamy J, Akondy RS, West EE et al. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8(+) T cells. Immunity 2011; 35: 400–412.
Gorelik G, Richardson B . Aberrant T cell ERK pathway signaling and chromatin structure in lupus. Autoimmun Rev 2009; 8: 196–198.
Gorelik G, Fang JY, Wu A, Sawalha AH, Richardson B . Impaired T cell protein kinase C delta activation decreases ERK pathway signaling in idiopathic and hydralazine-induced lupus. J Immunol 2007; 179: 5553–5563.
Deng C, Kaplan MJ, Yang J, Ray D, Zhang Z, McCune WJ et al. Decreased Ras-mitogen-activated protein kinase signaling may cause DNA hypomethylation in T lymphocytes from lupus patients. Arthritis Rheum 2001; 44: 397–407.
Ichiyama K, Chen T, Wang X, Yan X, Kim BS, Tanaka S et al. The methylcytosine dioxygenase tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity 2015; 42: 613–626.
Loots GG, Ovcharenko I, Pachter L, Dubchak I, Rubin EM . rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res 2002; 12: 832–839.
Acknowledgements
This work was supported by the Ministry of Science and Technology, Taiwan, ROC (MOST 103-2320-B-016-017-MY3 and MOST 104-2320-B-016-014-MY3 to H-KS; NSC 102-2321-B-016-005-MY3 to F-CC), and Tri-service General Hospital foundation (TSGH-C103-005-007-009-S01 and TSGH-C104-008-S02 to H-KS). We thank Drs Maja Klug and Michael Rehli for kindly providing us the CpG-free plasmids.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Genes and Immunity website
Supplementary information
Rights and permissions
About this article
Cite this article
Chou, FC., Kuo, CC., Chen, HY. et al. DNA demethylation of the TIM-3 promoter is critical for its stable expression on T cells. Genes Immun 17, 179–186 (2016). https://doi.org/10.1038/gene.2016.6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2016.6