Abstract
Cytokines expression can be influenced by polymorphisms in their respective coding genes. We associated the CTI/TTD haplotype (Hap-1) and TCI/CCI haplotype (Hap-2) in the IL4 gene formed by the −590, +33 and variable number of tandem repeat polymorphisms with the severity of chronic periodontitis in humans. The functionality of these IL4 haplotypes in the response of immune cells to phorbol 12-myristate 13-acetate (PMA) with Ionomycin and IL-1β (as inflammatory stimuli) was evaluated. Gene expression (quantitative real-time PCR), profile of secreted cytokines (multiplex) and phenotypic polarization of T cells (flow cytometry) were the outcomes assessed. Green fluorescent protein reporter plasmid constructs containing specific IL4 haplotype were transiently transfected into JM cells to assess the influence of the individual haplotypes on promoter activity. In response to inflammatory stimuli the immune cells from Hap-1 haplotype had increased expression of anti-inflammatory IL4; conversely, the Hap-2 haplotype showed higher levels of pro-inflammatory cytokines. The haplotype CTI proved to be the most important for the regulation of IL4 promoter, regardless of the nature of the inflammatory stimulation; whereas the polymorphism in the promoter region had the least functional effect. In conclusion, IL4 haplotypes studied are functional and trigger opposite immune responses: anti-inflammatory (Hap-1) and pro-inflammatory (Hap-2). In addition, we identified the CTI haplotype as the main responsible for the regulation of IL4 transcriptional activity.
Similar content being viewed by others
Introduction
DNA polymorphisms are ubiquitous genetic variations among individuals and include single-nucleotide polymorphisms (SNPs), insertions and deletions (indels), and other larger rearrangements.1 They differ from mutations because of their higher frequency in the general population (>1%). Gene polymorphisms may cause significant changes in function by altering the levels or activities of their specific proteins.2 The relationships between polymorphisms in immune system genes and multifactorial disease have been reviewed previously.3 Moreover, differences in cytokine levels have been attributed to polymorphisms in their respective genes, as demonstrated by interleukin-1 (IL-1),4 IL-10,5, 6 IL-6 (ref. 7) and IL-4.8
Interleukin 4 (IL-4) is a major immunomodulatory cytokine, mainly involved in adaptive immunity. It inhibits the secretion of pro-inflammatory cytokines such as IL-1, IL-6 and tumor necrosis factor (TNF), which potently downregulates macrophage function. Moreover, IL-4 acts as a mitogen of B cells and enhances the secretion of immunoglobulin G and immunoglobulin E (IgE),9 promoting humoral response associated with Th2-type responses.10 IL-4 is coded by the IL4 gene of ~10 kb in size and comprises four exons (GenBank accession no. M23442). The gene is located on chromosome 5q31.111 together with other Th2-related cytokine genes, such as IL-3, -5, -9, -13 and -15.12
Polymorphisms in the IL4 gene have been investigated and were associated with a variety of diseases characterized by modulation of the immune response, including asthma,13, 14 lupus erythaematosus,15 rheumatoid arthritis,16 preeclampsia,17, 18 coronary artery disease,19, 20 gastric cancer21, 22 and diabetes mellitus.23 Some of the most investigated polymorphisms in the IL4 gene are: the −590(T/C; promoter region, rs2243250), +33(C/T, 5′-untranslated region (UTR) or −34 counted from the start codon; rs2070874) and VNTR (variable number of tandem repeats; insertion (I)/deletion (D) of 70 base pairs in intron 3, or indel, rs2234665) (Figure 1a). However, the analysis of an individual polymorphism may not detect an association with a disease or clinical condition. In fact, we reported on the lack of association between the -590 (SNP) in the IL4 gene promoter and the prevalence of chronic periodontitis (CP) in a Brazilian population.24 Encompassing genetic analysis including various polymorphisms in linkage disequilibrium (LD) (instead of the study of an individual polymorphism) is more robust in the study of genetic basis for diseases.25 The three commonly studied polymorphisms in the IL4 gene are in physical proximity on the chromosome 5q31.1, composing haplotypes based on the LD among the polymorphisms. Strong LD was observed for example in an American population,26 while perfect LD was observed in Japanese27 and Dutch28 populations. When we investigated a larger Brazilian population focusing on IL4 polymorphisms, we found haplotypes,29 such as CTI (formed by -590C (first allele, 'C'), +33 T (second allele, 'T'), and the third allele by the Insertion of 70 base pairs ('I', 3 repetitions of 70 bp=the larger haplotype) of the Indel VNTR in intron 3). Considering the pair of homologous chromosomes, the haplotypes can be accessed as genotypes, for example, the haplotype CTI/TTD means that the CTI haplotype resides on one of the chromosome 5q31.1, while the TTD haplotype resides on its homologous chromosome. Our previous study of a Brazilian population29 identified these haplotypes showing the following LD in regard to the periodontal status ('healthy'/control or 'diseased'/CP): control group: −590 and +33 (D’=0.53, r2=0.25); −590 and indel (D’=0.73, r2=0.32); +33 and indel (D’=0.81, r2=0.34), and in the CP group: −590 and +33 (D’=0.66, r2=0.15); -590 and indel (D’=0.88, r2=0.14); +33 and Indel (D’=0.87, r2=0.39). Interestingly, we verified that Brazilian individuals carrying the haplotype TCI/CCI were five times more susceptible to CP (odds ratio (OR)adjusted=5.27, 95% confidence interval (CI)=2.28–12.18), whilst those carrying the CTI/TTD were markedly less susceptible to CP (OR)adjusted=0.29, 95% CI=0.08–0.88), after adjusting for confounding variables.29 These or other polymorphism combinations forming IL4 haplotypes have been investigated also in relation to CP30 and other diseases such as chronic obstructive pulmonary disease,28 common variable immunodeficiency,31 asthma32 and multiple sclerosis.26
Schematic representation of the IL4 gene and plasmid map (pAcGFP1-1 -Clontech). (a) IL4 gene with the following polymorphism positions: −590 (C/T) in the promoter, +33 (C/T) in the 5′-UTR (near exon 1, E1) and VNTR (indel) in the intron 3 (i3). (b) Map of the plasmid used for construction of the investigated IL4 haplotype. Synthesized 1.18 kb sequence corresponding to IL4 promoter (−1940 to +60 nucleotide) comprising the −590 and +33 SNPs was inserted upstream from the GFP reporter gene. In addition, a synthesized 2.6 kb sequence corresponding to the third intron of IL4 was inserted downstream of the reporter gene.
Besides identifying associations between genetic variations and various diseases/conditions, the investigation of the biological functionality of these haplotypes is of paramount relevance to the understanding of the biological mechanism and obtain insight into therapeutic/preventive intervention strategies. Nevertheless, studies demonstrating a functional role for genetic variations in the IL4 gene, as individual polymorphisms (for example, the −590 SNP33) or arranged as haplotypes;8, 34 have been influenced by different study designs. mRNA and protein levels are influenced both by cell type and stage of differentiation and also by the microenvironment.35
In the present study, we aimed to investigate the biological functionality of IL4 haplotypes CTI/TTD (Hap-1: −590[C/T], +33[T/T], intron 3 [insertion/deletion]) and TCI/CCI (Hap-2: −590[C/T], +33[C/C], intron 3 [insertion/insertion]), not because the LD among the polymorphisms, but because the previous significant finding of TCI/CCI were five times more susceptible to CP in a Brazilian population.29 The genetic results of that study29 demand to be biologically understood. In other words, functional comparisons between Hap-1 and Hap-2 compares [CT][TT][DI] with [CT][CC][II] and the differences between the groups could thus represent a difference between homozygous −33[C/C] versus −33[T/T] SNP or a difference between heterozygous indel in intron 3 or a homozygous insertion. We assessed the influence of these haplotypes in the response of immune cells to two distinct inflammatory stimuli, IL-1β and phorbol 12-myristate 13-acetate (PMA), verifying gene expression at mRNA and protein levels and immune cell polarization. We also determined the specific influence of each individual haplotype on IL4 gene promoter activity using gene reporter constructs.
Results
Subjects
Hap-1 or Hap-2 variations in the IL4 gene of the donors was confirmed by sequencing. There were no differences in age (Hap-1: 42.8±11.0 years standard deviation; Hap-2: 42.6±10.4) and gender (four female: two male in each group) between the six Hap-1 and six Hap-2 donors (P>0.05).
Influence on selected inflammatory gene expression according to the cell type
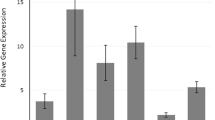
Initially, to provide some mechanistic insight into the functionality of the haplotypes on the response of individual immune cells to inflammatory stimuli, we assessed mRNA expression of IL4, IL8, IL12 and TNF-α by neutrophils, monocytes and lymphocytes separately, as shown in Figure 2. Expression of IL4 induced by both stimuli was significantly higher in the Hap-1 group compared with the Hap-2 group for all immune cell types: neutrophils (Figure 2a), monocytes (Figure 2b) and lymphocytes (Figure 2c). Conversely, expression of IL8 and TNFA was significantly higher in the Hap-2 group for all cell types (Figures 2a–c), whereas expression of IL12 was significantly greater in the Hap-2 group only in monocytes (Figure 2b).
Gene expression of IL4, IL8, IL12 and TNFA assessed in isolated cells from peripheral blood of individuals with the CTI/TTD IL4 (Hap-1) or TCI/CCI (Hap-2) haplotype after PMA+I or IL-1β stimuli. The mRNA levels (normalized by GAPDH) are represented in fold change in: (a) neutrophils; (b) monocytes; and (c) lymphocytes. Mean values of the relative gene quantifications of the Hap-1 and Hap-2 groups are represented in columns, and bars show standard deviations. *P<0.05, ** P<0.01.
Influence of IL4 haplotypes on cytokine production by peripheral immune cells
Cytokine production at the protein level was assessed in whole blood, in order to account for ‘global’ effect of the different haplotypes, including the interactions between the different immune cells and provide information that is more relevant from a perspective of the influence of these haplotypes on immune regulation. In the inflammatory panel, we observed significantly increased levels of pro-inflammatory cytokines IL-8 and TNF-α in the Hap-2 group. There were no differences in the GM-CSF levels, and higher levels of IL-1RA were observed in the Hap-1 group (Figure 3a). IL-4 was significantly greater in the Hap-1 group, while IFN-γ concentrations were higher in the Hap-2 group (Figure 3b). Also, significantly higher levels of IFN-α and IL-12 were observed in the Hap-2 group (Figure 3c). Regarding the chemokine panel, the Hap2- genotype was associated with significantly higher levels of Eotaxin, IP-10, MIG, MIP-1α and RANTES after stimulation with PMA/I (Figure 3d).
IL4 haplotytes and phenotypical polarization immune cells
There is a nonsignificant but noticeable trend towards a shift to the M1 profile in macrophages from individuals presenting the Hap-1 haplotype (Figures 4b and c). Monocytes from Hap-2 individuals showed significantly greater polarization towards the alternative M2 phenotype after stimulation (Figure 4d). On the other hand, T cells from individuals presenting Hap-2 haplotype, which was previously associated with susceptibility to CP, showed significant skewing towards pro-inflammatory Th1- and Th17-type responses (Figures 4f and i) and a concomitant significant inhibition of anti-inflammatory Th2- and Treg phenotypes (Figures 4g and j). These results indicate a contrasting influence of Hap-1 and Hap-2 haplotypes on the response of monocytes and T cells.
Cell phenotypic profile by flow cytometry from whole blood of individuals with the CTI/TTD IL4 (Hap-1) or TCI/CCI (Hap-2) haplotype after PMA+I stimulus. (a) Instrument set up and calibration. (b–d) M1 and M2 monocytes; (e–g) Th1 and Th2 lymphocytes; (h–j) Th17 and Treg lymphocytes. (a, e, i) Specific cell gate; (b, e, h) dot-blot representative of cells; (c, d, f, g, i, j) fold change calculated in comparison to the percentage of positively stained cells of the CTI/TTD haplotype in control/unstimulated conditions. **P<0.01.
Relative functional importance of the individual genetic variations forming the haplotypes in the IL4 gene
Different green fluorescent protein (GFP) reporter plasmid constructs, in which GFP expression was driven were: the (P) construct (containing only the promoter sequence of the IL4 gene, with T allele in the −590 SNP), as well the other constructs containing the different individual polymorphisms that form the studied haplotypes (TCI, TTD, (represented the Hap-1), CCI and CTI (represented the Hap-2). Each construct was transiently transfected by electroporation/cationic reagents (Neon system, Invitrogen Corp, Carlsbad, CA, USA) in JM cells (human T lymphocytes). These cells were stimulated with PMA+I, IL-1β, as inflammatory stimuli and also with IL-4 and anti-IL-12 to induce a Th2-type response. The GFP reporter gene expression was assessed after 24 h by flow cytometry. In general, we observed that (P) promoter sequence of IL4 was associated with the lowest expression of GFP reporter after all stimuli (Figures 5a–c). The CTI haplotype was the most important genetic variation, resulting in markedly increased expression of the GFP reporter gene in response to all stimuli (Figures 5a–c).
Transcriptional activity by GFP reporter gene of each IL4 haplotype construct in comparison with the construct containing only the promoter (P) region of the IL4 gene in which the T allele was present in the −590 SNP. JM cells were stimulated with: (a) PMA+I; (b) IL-4 and anti-IL-12; (c) IL-1β. **P<0.01; ***P<0.001.
Discussion
In this study, we describe the functionality of different haplotypes in the IL4 gene, which affects the response of immune cells to inflammatory stimuli and, thus, may be of diagnostic, prognostic and therapeutic relevance for chronic inflammatory conditions. The selected blood donors presenting the two haplotypes studied had similar age and gender. We investigated the role of two IL4 haplotypes formed by the −590(T/C), +33(C/T) and VNTR (indel) polymorphisms on gene expression and secretion of selected immune mediators upon inflammatory stimulation. We used an experimental approach that considered both the influence of IL4 haplotypes in different cell types individually (PMNs, monocytes, T cells) and on the net effect of interacting peripheral immune cells. Using GFP reporter plasmid constructs containing the different genetic variations that assemble into the distinct haplotypes in vivo, we determined the relative contribution of these genetic variations to IL4 promoter activity.
Nakashima et al.35 also analyzed the functionality of the same haplotypes on the IL4 gene in peripheral CD4+ T cells after stimulation with PMA+I. IL-4 gene expression was increased in CD4+ lymphocytes from individuals with the TTD/TTD haplotype. Our results also indicate increased IL-4 gene expression in individuals from a distinct racial background (Figure 2) and, moreover, we verified the functionality of this haplotype even in heterozygosity, as the donors in this study were heterozygous for this haplotype (CTI/TTD, Hap-1). Polymorphisms in the promoter region of IL4 gene can also be functional, as demonstrated by the increased IL4 and STAT6 mRNA levels, as well as IL-4 protein, in CD4+ cells of individuals with −590TT and −34TT IL4 genotypes.8 This finding agrees with the present study, as the individuals had the IL4 haplotype (−590)TT/(−34)TT8 (excluding the VNTR polymorphism), and our individuals had the CT/TT haplotype (part of Hap-1), which increased IL4 mRNA (Figure 2) and protein (Figure 3b). Another study on the functionality of IL4 haplotypes reported increased production of IFN-γ by peripheral blood mononuclear cells (PBMCs) of individuals homozygous for −590CC and VNTR-II.34 Considering the high reported LD between these polymorphisms, these individuals might have the C_I haplotype. In our study, we observed that the CCI haplotype was associated with higher production of IFN-γ, suggesting a common functional effect associated with a C_I haplotype. It is worth taking into mind that the present functional comparisons between Hap-1 and Hap-2 compares [CT][TT][DI] with [CT][CC][II], then the differences between the groups could represent here a difference between homozygous −33[C/C] versus −33[T/T]SNP or a difference between heterozygous indel in intron 3 or a homozygous insertion. The present transcriptional and post-transcriptional gene regulation findings associated with the investigated IL4 haplotypes could be due to not only the polymorphisms that form the referred haplotypes but also they could be occurring by the influence of a LD of the IL4 gene polymorphisms with others extending over longer distances to other genes of the 5q31 cytokine gene cluster, such as the IL-3/-4/-5/-9/-13/-15.26
The association of PMA and ionomycin used for the stimulation of cells is potent, polyclonal and nonspecific, which induces cytokine production. PMA is an analog of diacylglycerol, a key mediator of multiple intracellular signaling pathways.36 Ionomycin stimulates Ca2+ release from the endoplasmic reticulum, activating Ca2+-sensitive enzymes and synergizing with PMA.37, 38 IL-1β is the other 'inflammatory' stimulus used in this study, and engages IL-1R to trigger the expression of various inflammatory genes.39 The rationale for using these two different stimuli (PMA+ionomycin and IL-1 β) was to assess the functionality of IL4 haplotypes in the immune cells upon different activation pathways. Since the expression of target genes by immune cells was similarly affected by the two haplotypes after stimulation with PMA+I or IL-1β (Figures 2a–c); we performed subsequent experiments assessing the influence of IL4 haplotypes on protein production (flow cytometry and multiplex), using only the association PMA+I as the stimulus.
Most immune cells, including macrophages (CD14+ cells), neutrophils (CD15+ cells) and T-helper lymphocytes (CD3+/CD4+ cells) present functional/phenotypical heterogeneity, driven by a variety of microenvironmental cues. For example, IFN-γ, IL-1β and lipopolysaccharide induce the 'pro-inflammatory', classical phenotype of macrophage activation (M1), whereas IL-4 and IL-13 induce the 'anti-inflammatory/reparative' alternative phenotype of activation (M2).40 Similarly, neutrophils and CD4 T cells may be polarized into distinct 'pro-inflammatory' (Th1, Th17 for CD4+ cells, N1 for PMNs) or anti-inflammatory/reparative phenotypes (Th2, Tregs for CD4+ cells; N2 for PMNs).41, 42, 43, 44, 45, 46 Thus, variations on IL4 gene expression associated with the different haplotypes may have consequences for the phenotypical characteristics of immune cells.
Hap-2 haplotype is associated lower expression of IL4 and increased expression of IL8, IL12 and TNFA mRNA levels in neutrophils, monocytes and lymphocytes. Conversely, the same immune cell types isolated from donors presenting the Hap-1 haplotype show significantly greater expression of IL4 and reduced expression of IL-8, TNF-α and IL-12 (the latter is reduced only in monocytes). These results not only demonstrate that the haplotypes on IL4 gene are functional in terms of IL-4 expression but they also suggest that these haplotypes have a more 'global' effect on the immune response, represented by the skewing towards a general anti-inflammatory/reparative phenotype (M2/Th2/N2). These effects may be a direct result of the contrasting levels of IL-4 produced by cells from donors presenting the different haplotypes and influencing the microenvironment. Alternatively, differences in the patient’s genetic carriage (considering the IL4 haplotypes) could lead to chromatin changes at the signature cytokine loci (IFN-gamma/Th1 and IL-4/Th2), which could interfere at the level of Th1/Th2 differentiation.47 Future studies will investigate possible regulatory factors responsible for gene regulation.
Intriguingly, presence of the Hap-2 haplotype did not induce an M2 phenotype as assessed by flow cytometry (in contrast with a significant decrease on IL-12 mRNA expression by monocytes); however, this may be related with the fact that we used monocytes and not differentiated macrophages in these studies. Importantly, the analysis of secreted cytokines by the complex population of peripheral immune cells (experiments stimulating whole blood) supports this 'global' impact of the different IL4 gene haplotypes on the immune response, as cells from donors presenting Hap-1 haplotype had increased levels of anti-inflammatory cytokines (IL-1RA and IL-4), and cells from individuals presenting the Hap-2 haplotype showed increased levels of pro-inflammatory cytokines/chemokines (TNF-α, IL-8, GM-CSF, IL-2R, IFN-gamma, IFN-alpha, IL-12 (p40/p70), IL-15, eotaxin, IP-10, MIG, MCP-1, MIP-1alfa, MIP-1β and RANTES). This shows that Hap-2 haplotype is associated with an exacerbated inflammatory profile and, thus, may be of relevance in chronic or acute inflammatory conditions of infectious, autoimmune or even in the immune response to various types of cancer.48 In fact, presence of the CCI IL4 haplotype is associated with increased risk of multiple sclerosis.26 The same haplotype is investigated here as part of the Hap-2 haplotype (TCI/CCI).
Functional analysis of the relative influence of each specific haplotype (TCI, CCI, CTI and TTD) using GFP reporter expression showed that the CTI IL4 haplotype is the most relevant for upregulation of IL4 promoter activity. The CTI haplotype increased GFP expression by 14-fold (PMA+I), twofold (IL-4 and anti-IL-12) and 1.5-fold (IL-1B) in comparison with reporter construct including only the −590(T allele) in the promoter region (P). The variation in gene reporter expression with the distinct stimuli may suggest that the activation of different intracellular pathways and combination of DNA-binding protein (transcription factors/enhancer proteins/transcription repressors) factors. Alternatively, the different stimuli may induce production of other mediators that feedback on an autocrine loop to affect IL4 promoter activity. Future studies will look into the influence of the secreted proteins on IL4 promoter transcriptional activity of GFP in these constructs (or may be including polymorphisms in other genes of the 5q31 cytokine gene cluster), and also on the profile of DNA-binding proteins interacting with the IL4 promoter region. To our knowledge, this is the first study to use genetic constructs including intronic polymorphism VNTR (indel) to investigate the relative importance of distinct haplotypes on IL4 promoter activity.
Although the number of subjects included in this study allow for proper statistical inference, the verification of these results in samples of greater diversity of ethnicity will be relevant for the external validity of the information on the functionality of these IL4 gene haplotypes. Also, similarly to the fact that studying gene variations in haplotypes is more relevant than the study of SNPs; we cannot rule out that the IL4 gene haplotypes studied are accompanied by a myriad of other genetic variations (on the IL4 gene or other immune-related genes) that may also have a functional influence on the immune response. Nevertheless, the data presented has consistency and internal validity that supports a functional role for the haplotypes studied.
In summary, we show that the haplotypes studies are not only functional but also that these haplotypes have opposite effects on IL4 gene expression in PMNs, monocytes and T cells. Moreover, we expand these findings to demonstrate that these haplotypes on IL4 gene have a global effect in the phenotypical polarization of the immune response: prominently anti-inflammatory for Hap-1 and pro-inflammatory for Hap-2. Finally, we identify the CTI haplotype as the most relevant variation associated with increased IL4 promoter activity. This information will be expanded in future studies looking at the biological mechanisms mediating these functional effects of Hap-1 and Hap-2 haplotypes, which may provide insight into novel therapeutic strategies for conditions associated with dysregulated/exacerbated immune response. Identification of CTI haplotype as the most important genetic variation for the increased activity of IL4 promoter may be useful for risk analysis in disease prevention and treatment.
Materials and methods
Individuals
Individuals carrying the CTI/TTD haplotype (Hap-1; n=6) and TCI/CCI haplotype (Hap-2; n=6) IL4 gene were confirmed by sequencing. This study was approved by the Committee Ethical Affairs (CAAE 18527813.7.0000.5416). Sample size calculation was performed by the DDS Research (Sample Size Calculator, Average, two sample) utilizing IL4 gene expression values of samples from a pilot study. This calculation determined six subjects of each genotype with the ability to detect as significant (at the 80% confidence level) differences of 0.3 units on the averaged values with an estimated variation of 0.6 units. Utilizing the obtained data at the end of this study, power calculation analyses were performed also by the DDS Research, showing that for gene expression, protein concentration and flow cytometry, statistical power was greater than 80%. Therefore, a total sample size of six individuals was enough to have a sufficient power to detect differences between the groups for the evaluated data. Peripheral blood was collected by venipuncture always in the morning, with a fasting period of no less than 8 h. A total of five vacutainers (EDTA/K3-containing vacutainer tubes, Becton Dickinson Co., Franklin Lakes, NJ, USA), in a total volume of ~20 ml, were collected from each subject. All subjects were in good general health and did not make continuous use of any medication. Exclusion criteria were: use of antibiotics or of steroidal or non-steroidal anti-inflammatories in the previous past four months; current or past history of smoking, any autoimmune/allergic conditions, or any systemic disease with influence on the immune system, current pregnancy or lactation. The selected individuals consented to participate in this study, and a double-blinded researcher (G.A.) conducts the in vitro experiments.
Gene expression assay
Peripheral blood from the individuals was drawn into a vacutainer blood-collecting tube with EDTA/K3. Separation of nucleated cells was performed by a double gradient of Histopaque (Sigma, St Louis, MO, USA). After initial separation of neutrophils and PBMCs, monocytes and lymphocytes were isolated from other PBMCs using negative selection magnetic bead-based sorting (Dynabeads untouched human monocytes kit, #11350D; Dynabeads untouched human T cells kit, #11344D, Invitrogen). After separation, all cells were cultured overnight in RPMI 1640 supplemented with 1% heat-inactivated fetal bovine serum. Lymphocytes were activated with CD3/CD28 antibodies (Dynabeads Human T-Activator, #11131D, Invitrogen) for 7 h. After activation, concentration of cells was adjusted to 5 × 105 cells per ml for the 4 h stimulation (period established in a pilot study—data not shown) with with 50 ng ml−1 of PMA (Sigma) in addition to 500 ng ml−1 of ionomycin calcium salt (I) (Sigma) or 5 ng ml−1 of recombinant human interleukin-1β (IL-1 β; R & D Systems, Minneapolis, MN, USA) or medium only (control). Total RNA was extracted using an affinity column system including treatment to eliminate possible genomic DNA contaminants (RNAqueous kit, Ambion Inc., Austin, TX, USA). RNA was quantitated on a microvolume spectrophotometer (NanoView Plus, GE Healthcare, Munich, Germany) and 300 ng were used for cDNA synthesis using random hexamer primers and reverse transcriptase (High Capacity cDNA Reverse Transcription Kit, Applied Biosystems, Carlsbad, CA, USA). Real-time PCR was performed using TaqMan chemistry (Applied Biosystems) and pre-designed and optimized sets of primers and probe (Gene expression assays, Applied Biosystems) for detection of IL4 (NM_000589.2; cat#Hs00174122_m1), IL8 (NM_000584.3;cat#Hs00174103_m1), IL12A (NM_000882.3;cat#Hs01073447_m1), TNFA (NM_000594.2;cat#Hs01113624_g1). Expression of GAPDH (NM_002046.4;cat#Hs02758991_g1) was used as the endogenous control. Three independent experiments were performed in triplicate for each stimulus. The data was analyzed as relative changes to unstimulated controls by the DDCt method using the thermocycler’s software (StepOne Plus, Applied Biosystems). Since all experimental conditions were performed in aliquots of cells from every individual donor, regulation of target gene expression was determined as fold change over non-stimulated control for each donor, to account for individual variations in gene expression. The fold change values for each target gene in each of the six donors were then averaged according to the Hap-1 and Hap-2 genotype.
Cytokine production
Equal volumes of whole blood containing 1 × 106 PBMCs and RPMI 1640 supplemented with 20% heat-inactivated fetal bovine serum were combined and immediately stimulated PMA+Ionomycin for 12 h at 37 °C in a 5% CO2 atmosphere. Levels of cytokines were measured in whole blood using a bead-based multiplex assay (Human Cytokine 25-Plex Panel, cat#LHC0009, Invitrogen) utilizing the Bio-plex 200 (Bio-Rad, Hercules, CA, USA; Invitrogen), following the manufacturer’s instructions. The experiments were performed and analyzed in duplicate. Three independent experiments were performed in triplicate for each stimulus.
Immunophenotyping
Equal volumes of whole blood containing 1 × 106 PBMCs and RPMI 1640 supplemented with 20% heat-inactivated fetal bovine serum were combined and immediately stimulated with PMA (50 ng ml−1)+Ionomycin (500 ng ml−1) for 72 h at 37 °C in a 5% CO2 atmosphere. After stimulation, collection of samples included both non-attached cells, which were transferred to a tube, and the attached cells, which were detached with enzyme-free cell dissociation buffer (Gibco, Life Technologies, Carlsbad, CA, USA), and combined in the same tube containing the non-attached cells. Erythrocytes were lysed by incubation with FACS Lysing solution for 10 min (FACS Lysing Solution, BD Biosciences, San Jose, CA, USA). The leukocytes were collected by centrifugation (400 g, 5 min, RT), counted and adjusted to 1 × 106 cells per ml. These samples were then separated in three aliquots and two aliquots were stained for CD4/PeCy7 (T-helper cell marker; cat#348789, BD Biosciences) and the remaining aliquot stained for CD14/PeCy7 (monocyte marker; cat#557742, BD Biosciences) for 30 min in the dark. Cells in all three aliquots were permeabilized with saponin-containing buffer (Cytoperm, BD Biosciences) for 15 min. The two CD4-stained aliquots were subsequently stained for either IFNγ fluorescein isothiocyanate (FITC) (cat#552882)/IL-4 PE (cat#559333) or IL-17 pycoerythrin (PE) (cat#560436)/FoxP3 AlexaFluor 488 (cat#561181); whereas CD14-stained aliquots were stained for IL-12 FITC (cat#554574) and IL-10 PE (cat#554706). Staining was performed for 40 min in the dark according to the manufacturer’s instructions. Aliquots of these leukocytes were used for the preparation of experimentally stained samples, as well as the instrument controls, which included: unstained samples, single fluorophore-stained samples and isotype-control samples. These were all prepared following the same protocol, regarding dilution/staining buffer, number of washes and conditions of staining (concentration of antibodies and incubation period). Acquisition of data was always preceded by the verification of the performance of the flow cytometer (FACSVerse, BD Biosciences) using CS&T Research Beads (cat#650621, BD Biosciences), as recommended by the manufacturer. The acquisition strategy included setting up a side (SSC) × forward (FSC) scatter dot-blot, used to exclude cell debris. The first gate (P1) was set upon this plot to include all three main types of leukocytes (lymphocytes, monocytes and granulocytes) based on cell size and complexity. Two additional dot blots (FSC-Area × FSC-Width and SSC-Area × SSC-Width) were set up to exclude doublets and cell 'clumps', generating P2 (considering only 'P1' events, that is, leukocytes minus cell debris and excluding cell doublets) and P3 (considering only 'P2' events, that is, leukocytes and a 'second pass' to exclude cell doublets). A fourth dot plot (considering only 'P3' events) was set up with the parameters SSC-A × PE-Cy7 to identify both CD4+ and CD14+ cells, both stained by PE-Cy7-conjugated primary antibodies, generating 'P4' for CD4+ or CD14+ cells. Finally, a dot plot including the parameters FITC × PE (considering only 'P4' events) was used to assess Th1/Th2 (aliquot#1, IFNg/IL4) Treg/Th17 (aliquot#2, IL17A/FoxP3) and M1/M2 (aliquot#3, IL12/IL-10) using quadrant gates. Before data acquisition, unstained samples were used to set up the correct voltages of all photomultiplier tube. Then samples stained with a single antibody/fluorophore (PE-Cy7, PE and FITC) were used to adjust voltage of the photomultiplier tube for each channel and also the compensation between FITC and PE channels. Finally, aliquots of the samples stained with irrelevant isotype-matched control antibodies conjugated to the same fluorophores were used to verify false-positives due to nonspecific binding to Fc receptors. A total of 300.000 events were acquired for each experimental sample, and data was analyzed using the cytometer’s software (BD FACSuite, BD Biosciences). Three independent experiments were performed in triplicate for each stimulus.
Construction of IL4 haplotypes in GFP reporter vector
Plasmid GFP reporter vectors (pAcGFP1-1, Clontech Lab. Inc, cat#632497, Mountain View, CA, USA) were constructed by the GenScript Company such that the gene reporter expression (GFP) is under control of the proximal IL4 promoter (1.18 kb fragment, GenBank # M23442.1, comprising the −590 and +33 SNPs). Moreover, due to the importance of investigating the potential functionality of the VNTR (indel) polymorphism in the third intron of the IL4 gene, a 2.6 kb sequence corresponding to this intron was inserted immediately after the coding sequence of the GFP reporter gene vector and before the poly-A site of the GFP reporter vector (pAcGFP1-1, Clontech Lab. Inc.) (Figure 1b). Han et al. used similar construction in 201249 to investigate the transcriptional effect of an intronic polymorphism in the MYLK gene.
Site-directed mutagenesis
Two site-directed mutageneses were performed to obtain the CCI and CTI constructs (Table 1) from the original TCI plasmid construct synthesized by GenScript. Primers were designed for these site-directed mutations (SnapGene software, GSL Biotech, Chicago, IL, USA) (Table 1). The PCR for site-directed mutagenesis was performed with Phusion kit (New England Biolabs, Inc. Ipswich, MA, USA) and the polymerization reaction was performed in thermocycler (GeneAmp PCR System 9700 thermocycler, Applied Biosystems). The PCR product was transformed in competent Eshcherichia coli DH5α by thermal shock, and the plasmid purification was done with column-based Kit (QIAprep Spin Miniprep kit, Qiagen GmbH, Hilden, Germany) according to manufacturer's protocol. The plasmid DNA samples were quantitated in a spectrophotometer (NanoView Plus, GE Healthcare). The product was confirmed by sequencing (Big Dye Terminator kit v3.1 Cycle Sequencing Kit, Applied Biosystems). The quality and reliability of the sequenced products were checked (Sequence Scanner v 2.0 software, Applied Biosystems), and global multiple alignment was performed using the ClustalW2 and compared with the sequence according to the construct synthesized by GenScript (M23442 GenBank database). Thus, the plasmid containing the CCI mutation was used for site-directed mutagenesis reaction to obtain the CTI mutation.
Transfection and GFP reporter assays
The plasmid containing only the promoter sequence of the IL4 gene without the presence of SNPs +33(C/T) and VNTR (indel), referred to here as Promoter (P), the TCI and TTD plasmids (prepared by GenScript) and the CCI and CTI (mutagenized plasmids) were transiently transfected by combination of electroporation/cationic reagents (Neon system, Invitrogen) into the JM cells (human T lymphocytes), and the effects of the different haplotypes in the promoter and/or intron regions on the expression of GFP reporter gene were evaluated by flow cytometry. JM cells (2 × 107 cells per ml) were transfected with 10 μg of plasmid and the parameters used were: 1350 V, 10 ms and three pulses. After transfection, cells were incubated with RPMI 1640 medium supplemented with 10% fetal bovine serum for 24 h to recover. The cells were then stimulated for another 24 h with PMA (50 ng ml−1)/Ionomycin (500 ng ml−1), IL-1β (5 mg ml−1) and IL-4 (50ng ml−1; cat# 554605, BD Biosciences) with anti-IL-12 (1 mg ml−1; cat#554659, BD Biosciences) (to induce polarization towards the Th2 phenotype) and GFP expression was assessed by flow cytometry. Three independent experiments were performed in triplicate for each stimulus.
Statistical Analysis
To compare the outcomes of interest according to the IL4 haplotypes, analysis of variance and the unpaired t-test were performed utilizing the GraphPad Prism software (GraphPad Software, La Jolla, CA, USA). Differences were considered statistically significant at P<0.05.
Accession codes
References
Qi J, Chen Y, Copenhaver GP, Ma H . Detection of genomic variations and DNA polymorphisms and impact on analysis of meiotic recombination and genetic mapping. Proc Natl Acad Sci USA 2014; 111: 10007–10012.
Stern DL . Evolutionary biology. The problem of variation. Nature 2000; 408: 529–531.
Vandenbroeck K, Goris A . Cytokine gene polymorphisms in multifactorial diseases: gateways to novel targets for immunotherapy? Trends Pharmacol Sci 2003; 24: 284–289.
Shirodaria S, Smith J, McKay IJ, Kennett CN, Hughes FJ . Polymorphisms in the IL-1A gene are correlated with levels of interleukin-1alpha protein in gingival crevicular fluid of teeth with severe periodontal disease. J Dent Res 2000; 79: 1864–1869.
Turner D, Grant SC, Yonan N, Sheldon S, Dyer PA, Sinnott PJ et al. Cytokine gene polymorphism and heart transplant rejection. Transplantation 1997; 64: 776–779.
Crawley E, Woo P, Isenberg DA . Single nucleotide polymorphic haplotypes of the interleukin-10 5' flanking region are not associated with renal disease or serology in Caucasian patients with systemic lupus erythematosus. Arthritis Rheum 1999; 42: 2017–2018.
Smith AJ, D'Aiuto F, Palmen J, Cooper JA, Samuel J, Thompson S et al. Association of serum interleukin-6 concentration with a functional IL6 -6331 T>C polymorphism. Clin Chem 2008; 54: 841–850.
Gonzales JR, Groger S, Haley G, Bodeker RH, Meyle J . The interleukin-4 -34TT and -590TT genotype is correlated with increased expression and protein production in aggressive periodontitis. Mol Immunol 2010; 47: 701–705.
Tew J, Engel D, Mangan D . Polyclonal B-cell activation in periodontitis. J Periodontal Res 1989; 24: 225–241.
Mosmann TR, Coffman RL . TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol 1989; 7: 145–173.
Arai N, Nomura D, Villaret D, DeWaal Malefijt R, Seiki M, Yoshida M et al. Complete nucleotide sequence of the chromosomal gene for human IL-4 and its expression. J Immunol 1989; 142: 274–282.
Le Beau MM, Lemons RS, Espinosa R 3rd, Larson RA, Arai N, Rowley JD . Interleukin-4 and interleukin-5 map to human chromosome 5 in a region encoding growth factors and receptors and are deleted in myeloid leukemias with a del(5q). Blood 1989; 73: 647–650.
Zhang S, Li Y, Liu Y . Interleukin-4 -589C/T polymorphism is associated with increased pediatric asthma risk: a meta-analysis. Inflammation 2015; 38: 1207–1212.
Birbian N, Singh J, Jindal SK, Sobti RC . High risk association of IL-4 VNTR polymorphism with asthma in a North Indian population. Cytokine 2014; 66: 87–94.
Mahmoudi M, Tahghighi F, Ziaee V, Harsini S, Rezaei A, Soltani S et al. Interleukin-4 single nucleotide polymorphisms in juvenile systemic lupus erythematosus. Int J Immunogenet 2014; 41: 512–517.
Krabben A, Wilson AG, de Rooy DP, Zhernakova A, Brouwer E, Lindqvist E et al. Association of genetic variants in the IL4 and IL4R genes with the severity of joint damage in rheumatoid arthritis: a study in seven cohorts. Arthritis Rheum 2013; 65: 3051–3057.
Salimi S, Mohammadoo-Khorasani M, Yaghmaei M, Mokhtari M, Moossavi M . Possible association of IL-4 VNTR polymorphism with susceptibility to preeclampsia. Biomed Res Int 2014; 2014: 497031.
Gebhardt S, Bruiners N, Hillermann R . A novel exonic variant (221delT) in the LGALS13 gene encoding placental protein 13 (PP13) is associated with preterm labour in a low risk population. J Reprod Immunol 2009; 82: 166–173.
Basol N, Celik A, Karakus N, Ozturk SD, Yigit S . The evaluation of angiotensin-converting enzyme (ACE) gene I/D and IL-4 gene intron 3 VNTR polymorphisms in coronary artery disease. In Vivo 2014; 28: 983–987.
Seitz HK, Homann N . The role of acetaldehyde in alcohol-associated cancer of the gastrointestinal tract. Novartis Found Symp 2007; 285: 110–119 discussion 119-4, 198-9.
Sun Z, Cui Y, Jin X, Pei J . Association between IL-4 -590C>T polymorphism and gastric cancer risk. Tumour Biol 2014; 35: 1517–1521.
Bhayal AC, Krishnaveni D, Rao KP, Kumar AR, Jyothy A, Nallari P et al. Significant association of Interleukin4 Intron 3 VNTR Polymorphism with susceptibility to gastric cancer in a South Indian population from Telangana. PloS one 2015; 10: e0138442.
Basol N, Inanir A, Yigit S, Karakus N, Kaya SU . High association of IL-4 gene intron 3 VNTR polymorphism with diabetic peripheral neuropathy. J Mol Neurosci 2013; 51: 437–441.
Scarel-Caminaga RM, Trevilatto PC, Souza AP, Brito RB Jr., Line SR . Investigation of IL4 gene polymorphism in individuals with different levels of chronic periodontitis in a Brazilian population. J Clin Periodontol 2003; 30: 341–345.
Kinane DF, Hart TC . Genes and gene polymorphisms associated with periodontal disease. Crit Rev Oral Biol Med 2003; 14: 430–449.
Kantarci OH, Schaefer-Klein JL, Hebrink DD, Achenbach SJ, Atkinson EJ, McMurray CT et al. A population-based study of IL4 polymorphisms in multiple sclerosis. J Neuroimmunol 2003; 137: 134–139.
Suzuki I, Yamaguchi E, Hizawa N, Itoh A, Kawakami Y . A new polymorphism in the 5' flanking region of the human interleukin (IL)-4 gene. Immunogenetics 1999; 49: 738–739.
Van der Pouw Kraan TC, Kucukaycan M, Bakker AM, Baggen JM, van der Zee JS, Dentener MA et al. Chronic obstructive pulmonary disease is associated with the -1055 IL-13 promoter polymorphism. Genes Immun 2002; 3: 436–439.
Anovazzi G, Kim YJ, Viana AC, Curtis KM, Orrico SR, Cirelli JA et al. Polymorphisms and haplotypes in the interleukin-4 gene are associated with chronic periodontitis in a Brazilian population. J Periodontol 2010; 81: 392–402.
Holla LI, Fassmann A, Augustin P, Halabala T, Znojil V, Vanek J . The association of interleukin-4 haplotypes with chronic periodontitis in a Czech population. J Periodontol 2008; 79: 1927–1933.
Rezaei N, Aghamohammadi A, Mahmoudi M, Shakiba Y, Kardar GA, Mahmoudi M et al. Association of IL-4 and IL-10 gene promoter polymorphisms with common variable immunodeficiency. Immunobiology 2010; 215: 81–87.
Noguchi E, Shibasaki M, Arinami T, Takeda K, Yokouchi Y, Kawashima T et al. Association of asthma and the interleukin-4 promoter gene in Japanese. Clin Exp Allergy 1998; 28: 449–453.
Rosenwasser LJ, Klemm DJ, Dresback JK, Inamura H, Mascali JJ, Klinnert M et al. Promoter polymorphisms in the chromosome 5 gene cluster in asthma and atopy. Clin Exp Allergy 1995; 25 (Suppl 2) 74–78.
Bartova J, Linhartova PB, Podzimek S, Janatova T, Svobodova K, Fassmann A et al. The effect of IL-4 gene polymorphisms on cytokine production in patients with chronic periodontitis and in healthy controls. Mediators Inflamm 2014; 2014: 185757.
Nakashima H, Miyake K, Inoue Y, Shimizu S, Akahoshi M, Tanaka Y et al. Association between IL-4 genotype and IL-4 production in the Japanese population. Genes Immun 2002; 3: 107–109.
Matthews SA, Cantrell DA . New insights into the regulation and function of serine/threonine kinases in T lymphocytes. Immunol Rev 2009; 228: 241–252.
Chatila T, Silverman L, Miller R, Geha R . Mechanisms of T cell activation by the calcium ionophore ionomycin. J Immunol 1989; 143: 1283–1289.
Oh-hora M . Calcium signaling in the development and function of T-lineage cells. Immunol Rev 2009; 231: 210–224.
Weber A, Wasiliew P, Kracht M . Interleukin-1 (IL-1) pathway. Sci Signal 2010; 3: cm1.
Zhu L, Zhao Q, Yang T, Ding W, Zhao Y . Cellular metabolism and macrophage functional polarization. Int Rev Immunol 2015; 34: 82–100.
Jankovic D, Liu Z, Gause WC . Th1- and Th2-cell commitment during infectious disease: asymmetry in divergent pathways. Trends Immunol 2001; 22: 450–457.
Murphy KM, Reiner SL . The lineage decisions of helper T cells. Nat Rev Immunol 2002; 2: 933–944.
Weaver CT, Hatton RD . Interplay between the TH17 and TReg cell lineages: a (co-)evolutionary perspective. Nat Rev Immunol 2009; 9: 883–889.
Appay V, van Lier RA, Sallusto F, Roederer M . Phenotype and function of human T lymphocyte subsets: consensus and issues. Cytometry A 2008; 73: 975–983.
Neely CJ, Kartchner LB, Mendoza AE, Linz BM, Frelinger JA, Wolfgang MC et al. Flagellin treatment prevents increased susceptibility to systemic bacterial infection after injury by inhibiting anti-inflammatory IL-10+ IL-12- neutrophil polarization. PloS One 2014; 9: e85623.
Fridlender ZG, Sun J, Kim S, Kapoor V, Cheng G, Ling L et al. Polarization of tumor-associated neutrophil phenotype by TGF-beta: "N1" versus "N2" TAN. Cancer Cell 2009; 16: 183–194.
Fields PE, Kim ST, Flavell RA . Cutting edge: changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J Immunol 2002; 169: 647–650.
Damsker JM, Hansen AM, Caspi RR . Th1 and Th17 cells: adversaries and collaborators. Ann N Y Acad Sci 2010; 1183: 211–221.
Han YJ, Ma SF, Wade MS, Flores C, Garcia JG . An intronic MYLK variant associated with inflammatory lung disease regulates promoter activity of the smooth muscle myosin light chain kinase isoform. J Mol Med 2012; 90: 299–308.
Acknowledgements
This study was supported by State of São Paulo Foundation Research (FAPESP) grants #2013/17887-8 and #2014/04638-2.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Anovazzi, G., Medeiros, M., Pigossi, S. et al. Functionality and opposite roles of two interleukin 4 haplotypes in immune cells. Genes Immun 18, 33–41 (2017). https://doi.org/10.1038/gene.2016.47
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2016.47
This article is cited by
-
Interleukin-4 gene polymorphism (C33T) and the risk of the asthma: a meta-analysis based on 24 publications
BMC Medical Genetics (2020)
-
Cell death in cancer in the era of precision medicine
Genes & Immunity (2019)
-
Cell death pathologies: targeting death pathways and the immune system for cancer therapy
Genes & Immunity (2019)