Abstract
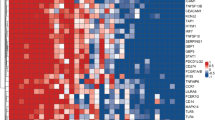
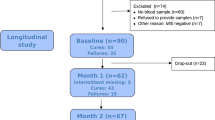
Tuberculosis (TB) is a major global health problem. Routine laboratory tests or newly developed molecular detection are limited to the quality of sputum sample. Here we selected genes specific to TB by a minimum redundancy–maximum relevancy package using publicly available microarray data and determine level of selected genes in blood collected from a Thai TB cohort of 40 active TB patients, 38 healthy controls and 18 previous TB patients using quantitative real-time PCR. FCGR1A, FCGR1B variant 1, FCGR1B variant 2, APOL1, GBP5, PSTPIP2, STAT1, KCNJ15, MAFB and KAZN had significantly higher expression level in active TB individuals as compared with healthy controls and previous TB cases (P<0.01). A mathematical method was applied to calculate TB predictive score, which contains the level of expression of seven genes and this score can identify active TB cases with 82.5% sensitivity and 100% specificity as compared with conventional culture confirmation. In addition, TB predictive scores in active TB patients were reduced to normal after completion of standard short-course therapy, which was mostly in concordant with the disease outcome. These finding suggested that blood gene expression measurement and TB Sick Score could have potential value in terms of diagnosis of TB and anti-TB treatment monitoring.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
O'Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, Berry MP . The immune response in tuberculosis. Ann Rev Immunol 2013; 31: 475–527.
Weyer K, Carai S, Nunn P . Viewpoint TB diagnostics: what does the world really need? J Infect Dis 2011; 204: S1196–S1202.
Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 2010; 466: 973–977.
Maertzdorf J, Repsilber D, Parida SK, Stanley K, Roberts T, Black G et al. Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun 2011; 12: 15–22.
Jacobsen M, Repsilber D, Gutschmidt A, Neher A, Feldmann K, Mollenkopf HJ et al. Candidate biomarkers for discrimination between infection and disease caused by Mycobacterium tuberculosis. J Mol Med 2007; 85: 613–621.
Streimish I, Bizzarro M, Northrup V, Wang C, Renna S, Koval N et al. Neutrophil CD64 as a diagnostic marker in neonatal sepsis. Pediatr Infect Dis J 2012; 31: 777–781.
Roussel M, Gros A, Sauvadet E, Gacouin A, Marque S, Chimot L et al. CD64, a reliable biomarker of bacterial infection in intensive care units? Am J Resp Crit Care Med 2012; 186: 1058.
Li S, Huang X, Chen Z, Zhong H, Peng Q, Deng Y et al. Neutrophil CD64 expression as a biomarker in the early diagnosis of bacterial infection: a meta-analysis. Int J Infect Dis 2012; 17: e12–e23.
Fjaertoft G, Hakansson L, Ewald U, Foucard T, Venge P . Neutrophils from term and preterm newborn infants express the high affinity Fcgamma-receptor I (CD64) during bacterial infections. Pediatr Res 1999; 45: 871–876.
Bloom CI, Graham CM, Berry MP, Wilkinson KA, Oni T, Rozakeas F et al. Detectable changes in the blood transcriptome are present after two weeks of antituberculosis therapy. PLoS One 2012; 7: e46191.
Mahasirimongkol S, Yanai H, Mushiroda T, Promphittayarat W, Wattanapokayakit S, Phromjai J et al. Genome-wide association studies of tuberculosis in Asians identify distinct at-risk locus for young tuberculosis. J Hum Genet 2012; 57: 363–367.
Mahasirimongkol S, Yanai H, Nishida N, Ridruechai C, Matsushita I, Ohashi J et al. Genome-wide SNP-based linkage analysis of tuberculosis in Thais. Genes Immun 2009; 10: 77–83.
Parida SK, Kaufmann SH . The quest for biomarkers in tuberculosis. Drug Discov Today 2010; 15: 148–157.
Anderson ST, Kaforou M, Brent AJ, Wright VJ, Banwell CM, Chagaluka G et al. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N Eng J Med 2014; 370: 1712–1723.
Bloom CI, Graham CM, Berry MP, Rozakeas F, Redford PS, Wang Y et al. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS One 2013; 8: e70630.
Cliff JM, Lee JS, Constantinou N, Cho JE, Clark TG, Ronacher K et al. Distinct phases of blood gene expression pattern through tuberculosis treatment reflect modulation of the humoral immune response. J Infect Dis 2013; 207: 18–29.
Jacobsen M, Mattow J, Repsilber D, Kaufmann SH . Novel strategies to identify biomarkers in tuberculosis. Biol Chem 2008; 389: 487–495.
Kaforou M, Wright VJ, Oni T, French N, Anderson ST, Bangani N et al. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study. PLoS Med 2013; 10: e1001538.
Maertzdorf J, Ota M, Repsilber D, Mollenkopf HJ, Weiner J, Hill PC et al. Functional correlations of pathogenesis-driven gene expression signatures in tuberculosis. PLoS One 2011; 6: e26938.
Sutherland JS, Loxton AG, Haks MC, Kassa D, Ambrose L, Lee JS et al. Differential gene expression of activating Fcgamma receptor classifies active tuberculosis regardless of human immunodeficiency virus status or ethnicity. Clin Microbiol Infect 2014; 20: O230–O238.
Siberil S, Dutertre CA, Boix C, Bonnin E, Menez R, Stura E et al. Molecular aspects of human FcgammaR interactions with IgG: functional and therapeutic consequences. Immunol Lett 2006; 106: 111–118.
van der Poel CE, Spaapen RM, van de Winkel JG, Leusen JH . Functional characteristics of the high affinity IgG receptor, FcgammaRI. J Immunol 2011; 186: 2699–2704.
Xie T, Liang J, Liu N, Wang Q, Li Y, Noble PW et al. MicroRNA-127 inhibits lung inflammation by targeting IgG Fcgamma receptor I. J Immunol 2012; 188: 2437–2444.
Serezani CH, Aronoff DM, Sitrin RG, Peters-Golden M . FcgammaRI ligation leads to a complex with BLT1 in lipid rafts that enhances rat lung macrophage antimicrobial functions. Blood 2009; 114: 3316–3324.
Ridruechai C, Mahasirimongkol S, Phromjai J, Yanai H, Nishida N, Matsushita I et al. Association analysis of susceptibility candidate region on chromosome 5q31 for tuberculosis. Genes Immun 2010; 11: 416–422.
Sarrazin S, Mossadegh-Keller N, Fukao T, Aziz A, Mourcin F, Vanhille L et al. MafB restricts M-CSF-dependent myeloid commitment divisions of hematopoietic stem cells. Cell 2009; 138: 300–313.
Kim H, Seed B . The transcription factor MafB antagonizes antiviral responses by blocking recruitment of coactivators to the transcription factor IRF3. Nat Immunol 2010; 11: 743–750.
Smith EE, Malik HS . The apolipoprotein L family of programmed cell death and immunity genes rapidly evolved in primates at discrete sites of host-pathogen interactions. Genome Res 2009; 19: 850–858.
Pays E, Vanhollebeke B . Human innate immunity against African trypanosomes. Curr Opin Immunol 2009; 21: 493–498.
Wasser WG, Tzur S, Wolday D, Adu D, Baumstein D, Rosset S et al. Population genetics of chronic kidney disease: the evolving story of APOL1. J Nephrol 2012; 25: 603–618.
Hartman SE, Bertone P, Nath AK, Royce TE, Gerstein M, Weissman S et al. Global changes in STAT target selection and transcription regulation upon interferon treatments. Genes Dev 2005; 19: 2953–2968.
Monajemi H, Fontijn RD, Pannekoek H, Horrevoets AJ . The apolipoprotein L gene cluster has emerged recently in evolution and is expressed in human vascular tissue. Genomics 2002; 79: 539–546.
Zhaorigetu S, Wan G, Kaini R, Jiang Z, ApoL1 Hu CA . a BH3-only lipid-binding protein, induces autophagic cell death. Autophagy 2008; 4: 1079–1082.
Ding C, Peng H . Minimum redundancy feature selection from microarray gene expression data. J Bioinform Comput Biol 2005; 3: 185–205.
Friedman J, Hastie T, Tibshirani R . Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010; 33: 1–22.
Acknowledgements
We thank all participants in this study. In addition, the authors are really appreciated all assistance in subject recruitment and all administrative works from the TB–HIV research foundation. This work was financially supported by the Department of Medical Sciences, the Ministry of Public Health, Thailand, the National Research Council of Thailand and the Japan Society for the Promotion of Sciences. The method for gene expression measurement described in this work is in the process of patenting in the Kingdom of Thailand (No. 1401004750).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Genes and Immunity website
Supplementary information
Rights and permissions
About this article
Cite this article
Satproedprai, N., Wichukchinda, N., Suphankong, S. et al. Diagnostic value of blood gene expression signatures in active tuberculosis in Thais: a pilot study. Genes Immun 16, 253–260 (2015). https://doi.org/10.1038/gene.2015.4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2015.4
This article is cited by
-
Integrative genomics of the mammalian alveolar macrophage response to intracellular mycobacteria
BMC Genomics (2021)
-
Multi-country evaluation of RISK6, a 6-gene blood transcriptomic signature, for tuberculosis diagnosis and treatment monitoring
Scientific Reports (2021)
-
A rare variant at 11p13 is associated with tuberculosis susceptibility in the Han Chinese population
Scientific Reports (2016)