Abstract
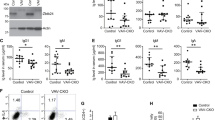
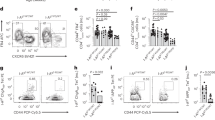
Invariant natural killer T (iNKT)-cell development is controlled by many polymorphic genes present in commonly used mouse inbred strains. Development of type 1 diabetes (T1D) in NOD mice partly results from their production of fewer iNKT-cells compared with non-autoimmune-prone control strains, including ICR. We previously identified several iNKT-cell quantitative trait genetic loci co-localized with known mouse and human T1D regions in a (NOD × ICR)F2 cross. To further dissect the mechanisms underlying the impaired iNKT-cell compartment in NOD mice, we carried out a series of bone marrow transplantation as well as additional genetic mapping studies. We found that impaired iNKT-cell development in NOD mice was mainly due to the inability of their double-positive (DP) thymocytes to efficiently select this T-cell population. Interestingly, we observed higher levels of CD1d expression by NOD than by ICR DP thymocytes. The genetic control of the inverse relationship between the CD1d expression level on DP thymocytes and the frequency of thymic iNKT-cells was further mapped to a region on chromosome 13 between 60.12 and 70.59 Mb. The NOD allele was found to promote CD1d expression and suppress iNKT-cell development. Our results indicate that genetically controlled physiological variation of CD1d expression levels modulates iNKT-cell development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Bendelac A, Savage PB, Teyton L . The biology of NKT cells. Annu Rev Immunol 2007; 25: 297–336.
Matsuda JL, Mallevaey T, Scott-Browne J, Gapin L . CD1d-restricted iNKT cells, the 'Swiss-Army knife' of the immune system. Curr Opin Immunol 2008; 20: 358–368.
Chen YG, Tsaih SW, Serreze DV . Genetic control of murine invariant natural killer T-cell development dynamically differs dependent on the examined tissue type. Genes Immun 2012; 13: 164–174.
Chan AC, Serwecinska L, Cochrane A, Harrison LC, Godfrey DI, Berzins SP . Immune characterization of an individual with an exceptionally high natural killer T cell frequency and her immediate family. Clin Exp Immunol 2009; 156: 238–245.
Montoya CJ, Pollard D, Martinson J, Kumari K, Wasserfall C, Mulder CB et al. Characterization of human invariant natural killer T subsets in health and disease using a novel invariant natural killer T cell-clonotypic monoclonal antibody, 6B11. Immunology 2007; 122: 1–14.
Kis J, Engelmann P, Farkas K, Richman G, Eck S, Lolley J et al. Reduced CD4+ subset and Th1 bias of the human iNKT cells in type 1 diabetes mellitus. J Leukoc Biol 2007; 81: 654–662.
Berzins SP, Cochrane AD, Pellicci DG, Smyth MJ, Godfrey DI . Limited correlation between human thymus and blood NKT cell content revealed by an ontogeny study of paired tissue samples. Eur J Immunol 2005; 35: 1399–1407.
Lee PT, Putnam A, Benlagha K, Teyton L, Gottlieb PA, Bendelac A . Testing the NKT cell hypothesis of human IDDM pathogenesis. J Clin Invest 2002; 110: 793–800.
Chen YG, Driver JP, Silveira PA, Serreze DV . Subcongenic analysis of genetic basis for impaired development of invariant NKT cells in NOD mice. Immunogenetics 2007; 59: 705–712.
Esteban LM, Tsoutsman T, Jordan MA, Roach D, Poulton LD, Brooks A et al. Genetic control of NKT cell numbers maps to major diabetes and lupus loci. J Immunol 2003; 171: 2873–2878.
Fletcher JM, Jordan MA, Snelgrove SL, Slattery RM, Dufour FD, Kyparissoudis K et al. Congenic analysis of the NKT cell control gene Nkt2 implicates the peroxisomal protein Pxmp4. J Immunol 2008; 181: 3400–3412.
Hammond KJ, Poulton LD, Palmisano LJ, Silveira PA, Godfrey DI, Baxter AG . alpha/beta-T cell receptor (TCR)+CD4-CD8- (NKT) thymocytes prevent insulin-dependent diabetes mellitus in nonobese diabetic (NOD)/Lt mice by the influence of interleukin (IL)-4 and/or IL-10. J Exp Med 1998; 187: 1047–1056.
Hong S, Wilson MT, Serizawa I, Wu L, Singh N, Naidenko OV et al. The natural killer T-cell ligand alpha-galactosylceramide prevents autoimmune diabetes in non-obese diabetic mice. Nat Med 2001; 7: 1052–1056.
Lehuen A, Lantz O, Beaudoin L, Laloux V, Carnaud C, Bendelac A et al. Overexpression of natural killer T cells protects Valpha14- Jalpha281 transgenic nonobese diabetic mice against diabetes. J Exp Med 1998; 188: 1831–1839.
Matsuki N, Stanic AK, Embers ME, Van Kaer L, Morel L, Joyce S . Genetic dissection of V alpha 14J alpha 18 natural T cell number and function in autoimmune-prone mice. J Immunol 2003; 170: 5429–5437.
Naumov YN, Bahjat KS, Gausling R, Abraham R, Exley MA, Koezuka Y et al. Activation of CD1d-restricted T cells protects NOD mice from developing diabetes by regulating dendritic cell subsets. Proc Natl Acad Sci USA 2001; 98: 13838–13843.
Sharif S, Arreaza GA, Zucker P, Mi QS, Sondhi J, Naidenko OV et al. Activation of natural killer T cells by alpha-galactosylceramide treatment prevents the onset and recurrence of autoimmune type 1 diabetes. Nat Med 2001; 7: 1057–1062.
Shi FD, Flodstrom M, Balasa B, Kim SH, Van Gunst K, Strominger JL et al. Germ line deletion of the CD1 locus exacerbates diabetes in the NOD mouse. Proc Natl Acad Sci USA 2001; 98: 6777–6782.
Ueno A, Wang J, Cheng L, Im JS, Shi Y, Porcelli SA et al. Enhanced early expansion and maturation of semi-invariant NK T cells inhibited autoimmune pathogenesis in congenic nonobese diabetic mice. J Immunol 2008; 181: 6789–6796.
Wang B, Geng YB, Wang CR ., CD1-restricted NK . T cells protect nonobese diabetic mice from developing diabetes. J Exp Med 2001; 194: 313–320.
Zekavat G, Mozaffari R, Arias VJ, Rostami SY, Badkerhanian A, Tenner AJ et al. A novel CD93 polymorphism in non-obese diabetic (NOD) and NZB/W F1 mice is linked to a CD4+ iNKT cell deficient state. Immunogenetics 2010; 62: 397–407.
Leiter E, Atkinson M (eds). NOD Mice and Related Strains: Research Applications in Diabetes, AIDS, Cancer and Other Diseases. R.G. Landes Company: Austin, TX, USA, 1998.
Tsaih SW, Khaja S, Ciecko AE, MacKinney E, Chen YG . Genetic control of murine invariant natural killer T cells maps to multiple type 1 diabetes regions. Genes Immun 2013; 14: 380–386.
Griewank K, Borowski C, Rietdijk S, Wang N, Julien A, Wei DG et al. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity 2007; 27: 751–762.
Hu T, Simmons A, Yuan J, Bender TP, Alberola-Ila J . The transcription factor c-Myb primes CD4+CD8+ immature thymocytes for selection into the iNKT lineage. Nat Immunol 2010; 11: 435–441.
Kageyama R, Cannons JL, Zhao F, Yusuf I, Lao C, Locci M et al. The receptor Ly108 functions as a SAP adaptor-dependent on-off switch for T cell help to B cells and NKT cell development. Immunity 2012; 36: 986–1002.
Dutta M, Kraus ZJ, Gomez-Rodriguez J, Hwang SH, Cannons JL, Cheng J et al. A role for Ly108 in the induction of promyelocytic zinc finger transcription factor in developing thymocytes. J Immunol 2013; 190: 2121–2128.
Sintes J, Cuenca M, Romero X, Bastos R, Terhorst C, Angulo A et al. Cutting edge: Ly9 (CD229), a SLAM family receptor, negatively regulates the development of thymic innate memory-like CD8+ T and invariant NKT cells. J Immunol 2013; 190: 21–26.
Jordan MA, Fletcher JM, Pellicci D, Baxter AG . Slamf1, the NKT cell control gene Nkt1. J Immunol 2007; 178: 1618–1627.
Jordan MA, Fletcher JM, Jose R, Chowdhury S, Gerlach N, Allison J et al. Role of SLAM in NKT cell development revealed by transgenic complementation in NOD mice. J Immunol 2011; 186: 3953–3965.
Bendelac A . Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med 1995; 182: 2091–2096.
Coles MC, Raulet DH . NK1.1+ T cells in the liver arise in the thymus and are selected by interactions with class I molecules on CD4+CD8+ cells. J Immunol 2000; 164: 2412–2418.
Schumann J, Pittoni P, Tonti E, Macdonald HR, Dellabona P, Casorati G . Targeted expression of human CD1d in transgenic mice reveals independent roles for thymocytes and thymic APCs in positive and negative selection of Valpha14i NKT cells. J Immunol 2005; 175: 7303–7310.
Wei DG, Lee H, Park SH, Beaudoin L, Teyton L, Lehuen A et al. Expansion and long-range differentiation of the NKT cell lineage in mice expressing CD1d exclusively on cortical thymocytes. J Exp Med 2005; 202: 239–248.
Zimmer MI, Colmone A, Felio K, Xu H, Ma A, Wang CR . A cell-type specific CD1d expression program modulates invariant NKT cell development and function. J Immunol 2006; 176: 1421–1430.
Weinreich MA, Odumade OA, Jameson SC, Hogquist KA . T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat Immunol 2010; 11: 709–716.
Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA . Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol 2013; 14: 1146–1154.
Honey K, Benlagha K, Beers C, Forbush K, Teyton L, Kleijmeer MJ et al. Thymocyte expression of cathepsin L is essential for NKT cell development. Nat Immunol 2002; 3: 1069–1074.
Borg ZD, Benoit PJ, Lilley GW, Aktan I, Chant A, DeVault VL et al. Polymorphisms in the CD1d promoter that regulate CD1d gene expression are associated with impaired NKT cell development. J Immunol 2014; 192: 189–199.
Zhang F, Liang ZY, Matsuki N, Van Kaer L, Joyce S, Wakeland EK et al. A murine locus on chromosome 18 controls NKT cell homeostasis and Th cell differentiation. J Immunol 2003; 171: 4613–4620.
Tsukamoto K, Ohtsuji M, Shiroiwa W, Lin Q, Nakamura K, Tsurui H et al. Aberrant genetic control of invariant TCR-bearing NKT cell function in New Zealand mouse strains: possible involvement in systemic lupus erythematosus pathogenesis. J Immunol 2008; 180: 4530–4539.
Chun T, Page MJ, Gapin L, Matsuda JL, Xu H, Nguyen H et al. CD1d-expressing dendritic cells but not thymic epithelial cells can mediate negative selection of NKT cells. J Exp Med 2003; 197: 907–918.
Wandstrat AE, Nguyen C, Limaye N, Chan AY, Subramanian S, Tian XH et al. Association of extensive polymorphisms in the SLAM/CD2 gene cluster with murine lupus. Immunity 2004; 21: 769–780.
Cox A, Ackert-Bicknell CL, Dumont BL, Ding Y, Bell JT, Brockmann GA et al. A new standard genetic map for the laboratory mouse. Genetics 2009; 182: 1335–1344.
Broman KW, Wu H, Sen S, Churchill GA . R/qtl: QTL mapping in experimental crosses. Bioinformatics 2003; 19: 889–890.
Sen S, Churchill GA . A statistical framework for quantitative trait mapping. Genetics 2001; 159: 371–387.
Wergedal JE, Ackert-Bicknell CL, Tsaih SW, Sheng MH, Li R, Mohan S et al. Femur mechanical properties in the F2 progeny of an NZB/B1NJ x RF/J cross are regulated predominantly by genetic loci that regulate bone geometry. J Bone Miner Res 2006; 21: 1256–1266.
Acknowledgements
We are grateful to the NIH tetramer core facility for providing us CD1d tetramers. This work was supported by the National Institutes of Health grants DK077443 and AI110963 (to Y-GC), DK46266 and DK95735 (to DVS), as well as by grants from the Helmsley Charitable Trust (2014PG-T1D048 to DVS), The Juvenile Diabetes Research Foundation, The American Diabetes Association and the Children’s Hospital of Wisconsin Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Genes and Immunity website
Supplementary information
Rights and permissions
About this article
Cite this article
Tsaih, SW., Presa, M., Khaja, S. et al. A locus on mouse chromosome 13 inversely regulates CD1d expression and the development of invariant natural killer T-cells. Genes Immun 16, 221–230 (2015). https://doi.org/10.1038/gene.2014.81
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2014.81
This article is cited by
-
Intrinsic factors and CD1d1 but not CD1d2 expression levels control invariant natural killer T cell subset differentiation
Nature Communications (2023)
-
Modulation of TCR signalling components occurs prior to positive selection and lineage commitment in iNKT cells
Scientific Reports (2021)
-
UBASH3A deficiency accelerates type 1 diabetes development and enhances salivary gland inflammation in NOD mice
Scientific Reports (2020)