Abstract
Germline variation at immunoglobulin (IG) loci is critical for pathogen-mediated immunity, but establishing complete haplotype sequences in these regions has been problematic because of complex sequence architecture and diploid source DNA. We sequenced BAC clones from the effectively haploid human hydatidiform mole cell line, CHM1htert, across the light chain IG loci, kappa (IGK) and lambda (IGL), creating single haplotype representations of these regions. The IGL haplotype generated here is 1.25 Mb of contiguous sequence, including four novel IGLV alleles, one novel IGLC allele, and an 11.9-kb insertion. The CH17 IGK haplotype consists of two 644 kb proximal and 466 kb distal contigs separated by a large gap of unknown size; these assemblies added 49 kb of unique sequence extending into this gap. Our analysis also resulted in the characterization of seven novel IGKV alleles and a 16.7-kb region exhibiting signatures of interlocus sequence exchange between distal and proximal IGKV gene clusters. Genetic diversity in IGK/IGL was compared with that of the IG heavy chain (IGH) locus within the same haploid genome, revealing threefold (IGK) and sixfold (IGL) higher diversity in the IGH locus, potentially associated with increased levels of segmental duplication and the telomeric location of IGH.
Similar content being viewed by others
Introduction
Immunoglobulins (IGs) or antibodies are essential components of the adaptive immune system that have key roles in processes associated with innate and adaptive immunity. They are expressed by B cells as either cell-surface receptors or secreted proteins, and are formed by two pairs of identical ‘heavy’ and ‘light’ kappa or lamda protein chains, encoded by genes located at three major loci in the human genome: the IG heavy (IGH) at 14q32.33, and the IG light lambda (IGL) and kappa (IGK) loci, located at 22q11.2 and 2p11.2, respectively.1 Specifically, through a unique mechanism referred to as V-(D)-J rearrangement,2 individual Variable (V), Diversity (D) and Joining (J) genes at the IGH locus, and V and J genes at either the IGK or IGL loci rearrange somatically at the DNA level to generate V-D-J and V-J regions that, after transcription and translation, encode the variable domains of the antibody.1 V-(D)-J rearrangement is accompanied by the random addition and deletion of nucleotides at the junctions of the combined V, D and J genes by terminal deoxynucleotide transferase. The extreme variability observed in expressed antigen-naïve B cell antibody repertoires is due to this combinatorial and junctional diversity, and partly ensures that the immune system is able to recognize and mount effective immune responses against a diverse range of potential pathogens. At the population and species level, IG haplotype and allelic variation also make important contributions to the diversity of expressed antibody repertoires;3, 4, 5, 6 however, the roles of IG genetic polymorphism in antibody function have not been comprehensively investigated.7
Over the past three decades, extensive catalogs of genetic polymorphisms have been established for human IGH, IGK and IGL genes (IMGT; the international ImMunoGeneTics information system, http://www.imgt.org).1,8 Importantly, not only has IG genetic diversity recently been implicated in inter-individual variation in expressed antibody repertoires,3, 4, 5 but genetic variants in both IG coding regions, as well as recombination signals (RSs) and regulatory sequences are also known to influence antibody expression and function, and mediate risk of disease phenotypes.9, 10, 11 However, our understanding of germline variability at the IGH, IGK and IGL loci remains severely limited, especially in terms of haplotype structure (that is, large segmental duplications (SDs) and deletions) as well as coding and non-coding sequence polymorphisms. We have begun to uncover this variability in the IGH locus, leading to complete nucleotide resolution descriptions of large structural variants (insertions, deletions, duplications and complex rearrangements), including novel functional IGHV genes and alleles.6
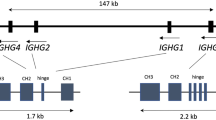
In the present study, we employ the same approach used for our previous characterization of IGH to analyze complete haploid reconstructions of the IGK and IGL loci, which to date, compared with IGH, remain less well investigated. The IGL locus (22q11.2) was initially mapped12, 13, 14 and fully sequenced and analyzed once in its entirety from multiple cosmid and BAC library resources (NG_000002.1).15 The locus spans approximately 0.9 Mb, and includes 69 IGLV, 7 IGLJ and 7 IGL constant (C) genes; IG gene alleles are designated as functional (F), open reading frame (ORF) or pseudogenes based on established criteria.1,16 Additional IGLV, IGLJ and IGLC genes not present in this haplotype are known to occur as insertion variants in the human population.1,17, 18, 19 Similarly, the initial mapping20, 21, 22 and sequencing of the IGK locus (2p11.2) was done using a composite of cosmid, bacteriophage and BAC clone libraries.23 A unique feature of the IGK locus is that it includes two large inverted SDs that comprise distinct V genes (termed proximal and distal); these regions remain separated by a large, currently unsequenced, assembly gap. The proximal region spans 0.54 Mb (NG_000834.1) and includes 69 IGKV functional/ORF genes and pseudogenes, whereas the distal region (NG_000833.1) spans 0.43 Mb and includes 62 V genes and pseudogenes (distal V genes are denoted by a ‘D’; for example, IGKV1D-13), five functional IGKJ genes, and a single functional IGKC gene reside downstream of the proximal V gene cluster.1 Additional BAC clones from the RPCI-11 library spanning the IGK proximal region and a portion of the distal region have also been sequenced.24 Both IGL and IGK are known to exhibit V, J and C gene allelic and structural variation.1,17, 18, 19,25, 26, 27
As a means to improve existing genomic resources in the IG loci, we have sequenced the IGK and IGL gene clusters from the CHORI-17 BAC library (CH17, BACPAC resources), previously constructed from a human hydatidiform mole cell line (CHM1htert). A hydatidiform mole occurs when a sperm fertilizes an enucleated egg. The paternal DNA is then doubled resulting in an effectively haploid genome. DNA resources from this material are thus ideal for resolving sequence assemblies in structurally complex genomic regions. Together with data produced previously for the IGH locus from the CH17 library,6 assemblies generated here for the IGK and IGL gene clusters represent the first complete haplotypes of all three IG loci from a single human. Using these data, we have conducted the first comparison of genetic diversity between the three IG regions in the same haploid genome. Our analyses have facilitated the identification of novel single-nucleotide polymorphisms (SNPs) within IGL and IGK gene coding regions, V gene RS sequences and regulatory elements, and also revealed evidence for a novel large sequence conversion event between the IGKV proximal and distal regions. In addition, our three-locus comparison shows a striking enrichment of structural and nucleotide diversity in the IGH locus, confirming previous suggestions that germline variation within the light chain gene loci is lower than that observed in IGH.18,28,29
Results
IGL and IGK reference sequences from the CH17 BAC library
We analyzed sequences from 17 CH17 BAC clones (IGL, 9; IGK, 8) comprising tiling paths across the two loci (Figures 1 and 2, Table 1)—one of the clones in IGK, CH17-158B1 (AC233264), had been sequenced previously. Clones unique to either IGK or IGL were then used to construct locus-wide contigs; clones in IGKV proximal and distal regions were aligned separately because the gap separating these two regions was not completely filled by the current sequencing effort.
IGL V, J and C gene comparison between CH17 and the NG_0000002.1 assembly. The upper panel shows the tiling path of sequenced CH17 BAC clones, with SD annotations depicted below. In the lower panel, functional and ORF IGL genes annotated in NG_0000002.1 and the CH17 haplotype are depicted by filled boxes, with corresponding locus and allele names located above and below the respective representation. Genes that are annotated as ORFs are denoted by green (V genes) or grey (J and C genes) boxes with a magenta outline. Shared genes and alleles between NG_0000002.1 and the CH17 haplotype are indicated by filled green or grey boxes. IGLV alleles that, in the CH17 haplotype, are different from those in NG_0000002.1 are indicated by boxes with other colors (red for non-synonymous and blue for synonymous allelic differences). IGLJ and IGLC allelic differences are denoted by the labelling of allele names on the CH17 haplotype. A filled purple circle denotes a novel allele in the CH17 haplotype with a polymorphism resulting in a non-sense mutation (pseudogene). All novel allele names are shown in bold. The 11-kb insertion in the CH17 IGL path is indicated.
IGK V, J and C gene comparison between CH17 and the NG_00034.1/NG000833.1 assemblies. The upper panel shows the tiling path of sequenced CH17 BAC clones, with SD annotations depicted below. The previously sequenced BAC clone, CH17-158B1, is noted by an asterisk. In the lower panel, functional or ORF IGK genes annotated in the NG_000834.1 (proximal) and NG_000833.1 (distal) assemblies and CH17 are depicted by filled boxes, with corresponding locus and allele names located above and below the respective representation. Genes that are annotated as ORFs are denoted by green (V genes) or grey (J and C genes) boxes with a magenta outline. IGKV genes and alleles that are shared between NG_000834.1/NG_000833.1 assemblies and CH17 are indicated by filled green boxes. IGKV alleles that, in the CH17 haplotype, are different from those in the NG_000833.2 (distal) and NG_000834.1 (proximal) assemblies are indicated by boxes with other colors (red for non-synonymous and blue for synonymous). Filled blue circles denote loci at which proximal or distal alleles were observed at the alternate locus (e.g., proximal allele at distal locus); and orange circles denote allelic differences between the haplotypes with respect to V-RSs. The 21-bp indel polymorphism upstream of IGKV1-8 is indicated with a red arrow. The novel sequence extending into the gap in the CH17 IGK path is shown in red. Horizontal black arrows indicate IGKV genes in opposite orientation in the locus, according to the IGK IMGT locus representation (IMGT Repertoire, http://www.imgt.org).1
For IGL, a single ~1.25 Mb contig was generated from nine BACs spanning 175.7 kb upstream of IGLV4-69 to 191.3 kb downstream of IGLC7 (Figure 1; GRCh37, chr22: 22209501–23456451). In total, we identified 37 of the 38 known functional/ORF IGLV genes, 7 functional J genes and 4 functional C genes. The remaining IGLV gene, IGLV5-39, was not found in CH17, consistent with it being an insertion polymorphism.1,18,27 Sequence comparisons within IGLV, IGLJ and IGLC genes revealed allelic differences between CH17 and the existing IGL assembly (NG_000002.1) at six V genes, two J genes and one C gene (Figure 1; Supplementary Table 1). Six of these alleles, five of which were novel (IGLV6-57*02, IGLV11-55*02, IGLV5-48*02, IGLV5-45*04 and IGLC7*03), included amino-acid changes. Notably, the novel allele identified at IGLV5-48 included a non-sense mutation that introduced a premature stop codon in the framework 3 region of the protein; 15 additional SNPs were also characterized within the exons of this gene. Before this study, only a single allele of IGLV5-48 had been described, classified as an ORF due to a single-nucleotide difference in the heptamer of the RS (V-RS TACAGTG instead of CACAGTG30). SNPs characterized in the remaining four novel functional/ORF alleles were represented in the 1000 genomes project (1KG) data set.31 Previously described regulatory motifs and RS sequences15 associated with each of the 37 identified functional/ORF IGLV genes were also inspected for previously uncharacterized variants in the CH17 haplotype, but no SNPs in these regions were identified.
Eight BAC clones were analyzed in the IGK locus, forming two independent contigs, one in the proximal region totalling 644 kb (GRCh37, chr2: 89027491–89630436), and a second in the distal region of the locus totalling 466 kb of contiguous sequence (GRCh37, chr2: 89841120–90308341; Figure 2). The proximal contig, containing four BACs, spanned from the intron of IGKV2-40 to 170 kb downstream of IGKC; thus, this contig lacked 10.9 kb of known sequence upstream of IGKV2-40 characterized in the initial genomic description of the locus (NG_000834.1), representing a small gap in the CH17 sequence. In the distal region, four complete BACs were assembled into a single contig spanning 22 kb upstream of the most distal gene IGKV3D-7 to 60 kb downstream of IGKV2D-40. This sequence included ~49 kb of additional sequence (compared with the originally sequenced distal cluster of IGK, NG_000833.1) extending into the assembly gap between the proximal and distal units, which is predicted to be 800 kb.23,32 The sequence extending into the unsequenced gap is dominated by complex repeats (Supplementary Figure 1), likely contributing to the difficulty of completing assemblies in this IG gene cluster. Alleles at 44 functional/ORF IGKV genes, 22 in each of the proximal and distal regions, as well as 5 IGKJ genes, and a single IGKC gene in the proximal region were characterized and compared with those found in the initial IGK assembly (NG_000834.1/NG_000833.1; Figure 2; Supplementary Table 2). To maintain consistency between IGL and IGK comparisons, for the analyses presented here we used IGK sequences initially generated by Kawasaki et al.23 (NG_000834.1/NG_000833.1) because existing GRCh37 assemblies also included a mixture of clones from the RPCI-11 BAC library, and a single clone from CH17 (CH17-158B1). On the basis of alignments of CH17 sequence to NG_000834.1/NG_000833.1, we observed 10 allelic differences of IGKV genes, and 1 allelic variant of IGKJ2; 10 of these alleles, including IGKJ2*04, involved amino-acid changes. We characterized 10 novel alleles that were not represented in IMGT, including 3 that were observed in NG_000834.1/NG_000833.1. In two instances, we observed the presence of alleles that had been previously classified as either ‘distal’ or ‘proximal’ alleles residing at loci in the alternate location. For example, we found a novel allele that matched with 100% sequence identity to IGKV1-13*02 at the IGKV1D-13 locus in the CH17 haplotype (this allele has been named IGKV1D-13*02). From our analysis of V-RS sequences, we found that in contrast to IGKV1D-13*01 in NG_000833.1, the allele in the CH17 haplotype at this locus was associated with a canonical, non-mutated V-heptamer sequence. Also, genomic descriptions of the gene IGKV1-8 have revealed a 21-bp deletion in an upstream regulatory element, which had been predicted to disrupt promoter function and inhibit expression (mutation observed in NG_000834.1);33 however, IGKV1-8*01 has been shown to be expressed in some cases (documented in IMGT Gene tables (www.imgt.org).34 Potentially explaining this discrepancy, we found that the previously described IGKV1-8*01 21 bp promoter deletion was not present in the CH17 haplotype, suggesting that this germline indel variant could contribute to variation in the expression of alleles at this locus.
Characterization of structural variants in IGL and IGK CH17 haplotypes
A direct comparison of the IGL CH17 and NG_000002.1 revealed the presence of only a single structural variant. This 11.9-kb insertion was located in the region between the pseudogenes IGLV7-35 and IGLV2-34 within the BAC, CH17-242N13; the region between these pseudogenes spans ~120 kb and is devoid of IG genes. Gene prediction analysis did not identify any genes within the insertion, nor did the insertion disrupt the non-IG related genes, ZNF280B, ZNF280A and PRAME, located in this region; the breakpoints of the event occurred between ZNF280A and PRAME. No structural variants were observed in the IGK CH17 haplotype.
We also searched the CH17 haplotypes for all IGL and IGK gene/allele sequences classified as ‘not located’ in the IMGT database/GENE-DB35 and IMGT Gene tables (www.imgt.org), meaning that these genes have not been located within IGL or IGK loci. Using this approach we mapped the pseudogene IGLV2-NL1 to the locus in CH17 corresponding to the position of the pseudogene IGLV2-34 in NG_000002.1; IGLV2-NL1 matched CH17 at this locus with 100% sequence identity, confirming that IGLV2-NL1 and IGLV2-34 genes are allelic rather than distinct loci. No additional exact matches between the CH17 haplotype and other ‘not-located’ IGKV or IGLV genes were observed.
Analysis of proximal and distal regions in CH17 haplotype and NG_000834.1/NG_000833.1 assemblies reveals evidence for sequence exchange
Previous analysis of sequence similarity between shared homology blocks of proximal and distal SD units, which comprise the majority of the IGK locus, revealed that over the majority of the locus these two clusters are >98% similar.23 SDs are known to facilitate sequence exchange via non-allelic homologous recombination and interlocus gene conversion;36,37 however, given the lack of reference sequence data this has not been investigated in the IGK locus. To address this, we conducted pair-wise comparisons of distal and proximal regions from CH17 and NG_000834.1/NG_000833.1 assemblies to search for large tracts of shared sequence (Figure 3a). The expectation is that sequences should be most similar between homologous regions of the NG_000834.1/NG_000833.1 and CH17 assemblies, whereas higher similarity between proximal and distal regions within a given assembly would suggest the potential occurrence of sequence exchange. Using this approach, we identified a large ~16.7-kb region that showed higher identity between the proximal and distal units of the NG_000834.1/NG_000833.1 assemblies than between the CH17 distal and NG_000834.1 IGK proximal units (Figure 3a). This region included two IGKV genes for which we observed allelic variants between the NG_000834.1/NG_000833.1 assemblies and CH17 haplotypes. Four-way sequence alignments of this region show that the CH17 distal unit was most unique compared with the other three sequences (Figure 3b), providing evidence that distal and proximal units have undergone sequence exchange at some point in the past, making the distal unit more similar to the proximal unit in this region represented in the NG_000834.1/NG_000833.1 assemblies. It is important to note, however, that the distal fragment of the initial assembly generated by Kawasaki et al.23 (NG_000833.1) harbors many unique bp differences compared with the other three sequences (blue tick marks, Figure 3b), which could be suggestive of the occurrence of mutation following the predicted sequence exchange event. Further analysis of this multi-sequence alignment using the Difference of Sums of Squares (DSS) method for recombination detection also predicted two potential flanking recombination breakpoints within the expected regions based on visual inspection of the sequence alignment and comparison of sequence similarities. We also analyzed sequence from two BAC clones in the proximal and distal clusters from the RPCI-11 BAC library;24 providing further support for this predicted event, this analysis revealed that these carried the same variants observed in the NG_000834.1/NG_000833.1 assemblies. An alternative explanation, although less parsimonious given the data presented, could be that this region of the IGKV distal cluster in the CH17 haplotype has undergone more rapid sequence divergence since the original duplication of the IGKV distal and proximal segments. The sequencing of additional haplotypes in the distal and proximal IGKV clusters will likely provide more insight into the mechanisms underlying these observed sequence signatures.
Detection of putative sequence exchange event between IGK proximal and distal gene clusters. (a) Pair-wise alignments between proximal and distal SDs in the CH17 haplotype and NG_000834.1 (proximal) and NG_000833.1 (distal) assemblies (Abbreviations: Ka, NG_000834.1/NG_000833.1 assemblies; prox, proximal; dist, distal). The region where Ka-prox and Ka-dist show stronger similarity than CH17-dist and Ka-prox highlights a potential region of sequence exchange between the two NG_000834.1 and NG_000833.1 assemblies (red box). Coordinates (GRCh37) for the proximal cluster are shown on the X axis. (b) Top panel shows a four-way sequence alignment of a 22.5-kb region from the proximal and distal units from within the red box in (a). Blue tick marks indicate bp SNP differences between the sequences. Upward pointing red arrows indicate boundaries of regions where the NG_000833.1 distal sequence aligns with a higher sequence similarity to the NG_000834.1/NG_000833.1 assemblies and CH17 proximal sequences than to the CH17 distal sequence, indicative of exchange between proximal and distal regions of the NG_000834.1/NG_000833.1 assemblies (sequence similarities: Ka-dist/CH17-dist=98.7%; Ka-dist/Ka-prox=99.7%). A DSS recombination analysis (McGuire and Wright;70 see Materials and Methods) using the same four-way sequence alignment is shown in the bottom panel. The two peaks with the strongest DSS values (downward pointing red arrows) correspond to the predicted breakpoints shown in the top panel based on sequence similarity values. The dotted line across the chart indicates the significance threshold based on the null distribution of DSS values calculated assuming no recombination.
SNP diversity and genomic features in IGHV, IGLV and IGKV gene regions
Compared with the number of alleles that differ between the IGHV CH17 haplotype and the GRCh37 assembly38 (NG_001019.5; 19 allelic variants/40 IGHV genes), V gene allelic variation described in this study for IGL (6 alleles/37 IGLV genes) and IGK (10 alleles/44 IGKV gene) was noticeably lower. This prompted us to also compare other genomic characteristics between the three loci. Excluding regions of structural variation between haplotypes/assemblies, we first generated SNP calls (not including gaps and single bp indels) between the CH17 and reference assemblies for all three loci; 519, 854 and 2897 SNPs were identified for IGKV, IGLV and IGHV, respectively (Table 2; Figure 4, left panel). After cross referencing these SNPs with dbSNP135 and 1KG data sets, 85, 103 and 407 SNPs in the IGKV, IGLV and IGHV loci were determined to be novel variants, not represented in either data set. Not surprisingly, given the number of SNPs in the 1KG data sets, fewer SNPs at each locus were represented in dbSNP (Figure 4). We examined these sites in publicly available Illumina data generated from the CHM1 genomic DNA39 to determine whether the novel SNPs were supported by an orthogonal platform (see Materials and Methods). We identified 84/85, 96/103 and 406/407 sites in IGKV, IGLV and IGHV, respectively, that are supported by the Illumina data. The discrepancies may represent sequencing errors. The novel SNPs for each region are reported in GRCh37 coordinates in Supplementary Table 3.
Representation of SNPs identified in the CH17 IG V gene regions in public SNP databases. The number of SNPs identified in IGKV, IGLV and IGHV gene regions based on alignments of CH17 to assemblies generated by Kawasaki et al.15,23 and Matsuda et al.38 (excluding gaps; NG_000834.1, IGK proximal; NG_000833.1. IGK distal; NG_000002.1, IGL; NG_001019.5 IGH). The fraction of SNPs represented in dbSNP135 (top) and 1KG (bottom) databases (db) is shown, and the total number of novel SNPs not found in either database is indicated at the bottom of the left panel.
Consistent with observations based on V gene allelic variation, SNP density in IGHV (0.0035) was approximately threefold higher than in IGLV (0.0012) and sixfold higher than in IGKV (0.0006); SNP densities were slightly elevated within functional/ORF V genes in each region compared with the calculated locus-wide values (Table 2). An analysis of genomic features in the three loci also showed that the fraction of each locus covered by repeat content was highest in IGHV, whereas that covered by SDs, not surprisingly was highest in IGKV (Table 2). However, when the primary SDs associated with the IGKV proximal/distal duplication event were excluded (that is, we only included SD annotations corresponding to duplications occurring solely within either proximal or distal clusters, but not between the two), SD coverage in IGHV (37.7%) was found to be much higher than both of the light chain loci (IGLV, 24.6%; IGKV, 28.2%; Table 2). When only SDs exhibiting >95% sequence identity were considered, this difference was even more striking (IGHV, 28.8%; IGLV, 12.9%; IGKV, 1.0%). Interestingly, in IGHV, SNP density was higher in regions of SD compared with regions not covered by SDs, especially true when considering only those regions covered by SDs with >95% sequence identity (Table 2); however, this difference was not found to be significant after permuting SD vs non-SD region assignments and re-assessing SNP densities of 10 000 permutations using the MCFDR (Monte Carlo False Discovery Rate) method40 (P>0.05). SNP density was also increased in segmentally duplicated regions of IGKV with >95% sequence identity compared with regions not covered by SDs, but again, this difference was not significant after permutation (P>0.05).
Given the telomeric location of IGHV and the known differences in nucleotide substitution patterns within telomeres compared with the rest of the genome,41 we next assessed CHM1 SNP density within and around autosomal telomeres and centromeres, again using variant data called from Illumina sequence of CHM1 genomic DNA.39 We found that mean SNP densities were ~2-fold higher in telomeric regions (within 3 Mb=0.001; 1 Mb=0.0009) when compared with centromeric regions (within 3 Mb=0.0005; 1 Mb=0.0004; Supplementary Figure 2A). Interestingly, SNP density within 1 Mb of the q arm telomere of chr14 harboring IGHV (0.002) was higher than all other telomeric regions, >2-fold higher than the telomeric average (Supplementary Figure 2B). In contrast, when CHM1 SNP densities within 3 Mb of all telomeres were compared, the region on the q arm of chr14 (including both IGHV and non-IGHV sequence) no longer stands out, suggesting that IGHV may have some unique properties contributing to higher than average genetic diversity. To place this in the context of the analysis conducted within the IG regions above, we also found that the q arm of chr14 has the second highest overlap with SDs (34%) in the genome (Supplementary Figure 3).
Discussion
We present data for the first haplotypes of the human IGL and IGK loci from the same haploid genome, representing only the second full-length assemblies constructed for these regions to date. From the CH17 clones, thirteen novel alleles were identified in the two loci, including four IGLV alleles, eight IGKV alleles, and one IGLC allele; an additional three IGKV alleles were also described from NG_000834.1/NG_000833.1 as part of our analysis. Two recent assessments of IGK alleles—one of a public data set of 435 expressed sequences,42 and a second of deep-sequenced antibody repertoires from four individuals43—concluded that, unlike IGH, IGK allelic data sets are likely to be mostly complete, as only two putative novel alleles were identified from these analyses.42 In contrast, IGHV sequencing in 10 individuals also supports these observations, finding that nearly 25% of characterized putative alleles were novel.44 However, the fact that we identified 12 novel light chain V gene alleles from a single haploid genome implies that additional efforts to identify unreported alleles in IGL and IGK are warranted. Importantly, as noted previously in IGHV,6 SNPs associated with novel alleles identified in IGL and IGK were present in the 1KG data set, further supporting the notion that the 1KG data set could serve as a useful resource for future investigations of IG gene diversity and the identification of novel polymorphisms. However, given the prevalence of SD in the IG loci, it will undoubtedly be essential to consider the impact of paralogous sequence variants when assessing 1KG SNP data in these regions, as complex and duplicated sequence structure is known to confound SNP characterization.45,46
In addition to novel allelic variants within IGL and IGK coding regions, variants involving an RS and regulatory element of two IGKV genes (IGKV1-8 and IGKV1D-13) were also identified. In both cases, RS and promoter sequences previously associated with alleles at these loci were predicted to inhibit their expression; however, the alleles described for these genes in the CH17 haplotype included variants that would be expected to exhibit normal gene expression. The importance of such polymorphisms is that they can result in variable levels of gene expression, including the loss of genes/alleles from expressed repertoires. In addition to those noted above, several other IGKV genes are also classified as ORF genes based on irregular RSs.34 Notably, IGKV2D-29*02, previously referred to as ‘A2c’, has a non-canonical V-heptamer, but has also been shown to occur in productive rearrangements. A third allele at the IGKV2D-29 locus, ‘A2b’, has also been shown to have a defective RS V-heptamer, which results in decreased expression and has been implicated in susceptibility to Haemophilus influenzae type b infection.9,47
Perhaps the most important contribution of the full IGK and IGL sequences of the CH17 haplotype data presented here is that, for the first time, locus-wide genetic diversity between IGH, IGL and IGK could be compared in the same haploid genome in relation to corresponding reference assemblies. Our first observation from this comparison was that the number of V gene allelic differences among the CH17 haplotypes was highest in IGHV. This finding is perhaps not surprising given that allelic richness is also known to be highest in IGHV based on available genetic data in the IMGT/GENE-DB35 and IMGT Allele Alignments (www.imgt.org). In addition, studies of substitution patterns in V genes have also revealed evidence for increased diversity in IGHV compared with IGLV and IGKV.29,48 Strikingly, however, when we extended our comparison to include all SNPs within the three loci, we found that locus-wide genetic diversity was also much higher in IGHV, indicating that increased diversity in this locus is not limited to within V gene coding regions. IGHV SNP density is also higher than that observed for killer cell IG-like receptor/leukocyte Ig-like receptor and T-cell receptor alpha loci, calculated at ~0.0016 for both loci (Steinberg et al., manuscript in preparation).
Similar to patterns noted from V gene allelic diversity and substitution patterns, fewer copy number variants (CNVs) have also been reported in IGLV and IGKV compared with IGHV. In IGL, for example, only three insertion-deletion variants of functional genes have been identified, involving IGLV1-50, IGLV8-61 and IGLV5-39;18,27 however, the deletion of IGLV8-61 has been identified in only a single individual. Likewise, in the IGKV locus, aside from an identified rare haplotype containing a deletion of the entire distal IGKV gene cluster,49,50 only a single functional V gene insertion, including the gene IGKV1-NL1, has been identified.25 Limited evidence suggesting putative IGKV gene duplications/deletions (for example, IGKV1-5 and IGKV3-20) has also more recently been noted from expressed antibody repertoire data, although these await confirmation as germline polymorphisms.43 Thus, in total, liberal estimates from the literature indicate that only 4–6 light chain functional/ORF V genes are known to occur in CNV, in stark contrast to ~29 in IGHV.6,7 In line with this difference, four V gene-containing CNVs were identified in the CH17 IGHV haplotype compared with zero defined in IGLV and IGKV.
Two factors suggest that the higher rate of CNV in IGH may be attributable to the increased fraction of the locus covered by segmentally duplicated sequences. First, SDs are known to be associated with SVs genome -wide,51 and second, duplications have been shown specifically to facilitate structural variation in IGH.6 SDs and tandem repeats also mediate sequence exchange either through gene conversion or recombination, that can result either in the homogenization of paralogous sequences,36,37,52 or in an increase in genetic diversity.53,54 Illustrating the latter of these two scenarios, we found that for IGHV, SNP density was highest in regions including SDs, particularly those with >95% sequence identity with their paralogs; similar trends were not noted for SNP density estimates calculated within non-SD repeat regions (Table 2). Importantly, SDs with >95% identity comprised nearly twice the fraction of IGHV sequence than IGLV sequence, which may in part explain the differences observed in SNP density between IGH and IGL. It is also worth noting that the difference in the fraction of SDs between IGH and the light chain regions would be greater if the 222 kb of novel sequence (comprised primarily of SDs) identified in IGH by our previous study6 was also included. In addition to this, assessments of CHM1 SNP density across autosomal telomeres and centromeres revealed that the telomeric region on chr14 containing IGHV had both elevated levels of SNP diversity, and increased SD overlap, compared with analogous regions on other autosomes. This suggests that the genomic location of IGHV has also likely contributed to the increased genetic variation we observe in this locus compared with IGKV and IGLV.
The distinct clustering and genomic partitioning of V subgroups12 within the IGL locus, and the observation that, compared with IGH and IGK, there are fewer IGL orphons present in other regions of the genome, has prompted the suggestion that IGL genes have undergone less ‘evolutionary shuffling’,28 which may be linked to lower levels of diversity in the locus18 and would be consistent with the results presented here. In comparison with the other two loci, we found IGKV to have the lowest locus-wide SNP density, nearly sixfold lower than that observed in IGHV. Due to the large inverted duplication of the proximal and distal regions, over 80% of the IGKV locus consists of SDs with >95% sequence identity. This suggests that, unlike in IGHV, SDs may be responsible for sequence homogenization rather than an increase in SNP diversity. The fact that we found evidence of a large tract of sequence exchange between the proximal and distal IGKV units lends support to this notion. However, fully understanding the relationship between SD and SNP density in the human IG regions will undoubtedly require further sequencing and comparisons of additional haplotypes. We must also acknowledge the potential for confounding effects related to the use of mosaic reference sequences for this comparison,15,23,38 which were generated from multiple large insert libraries constructed from diploid tissues from different individuals of different, or in some cases of unknown, ethnicity. However, because our findings are supported by existing V gene allelic variation data at the population level, it seems unlikely that the difference in variability between loci is due to the ethnic origin of the tissue.
If the difference in SNP density observed here between the IG V gene clusters is in fact genuine, then it raises the question of whether increased genetic diversity in IGH has any functional consequences. Given that SNP density within V gene coding regions in the CH17 haplotype was also higher in IGH compared with IGL and IGK, it could be speculated that mechanisms associated with an increased number of polymorphisms locus wide in IGHV, could by default, result in greater IGHV gene diversity and a more variable expressed antibody repertoire. Intriguingly, in natural antibodies, the IG heavy chains are considered to have a more prominent role in epitope binding than IG light chains, although this is primarily attributed to the third complementarity determining region (VH CDRH3) not encoded by IGHV genes.55 However, there are several examples demonstrating essential functions of IGHV germline-encoded variation in antigen specificity; for example, amino acids encoded by germline IGHV1-69 alleles have been shown to make important contributions to neutralizing antibody responses against influenza, hepatitis C and Middle East Respiratory Syndrome coronavirus.10,56,57 A recent mapping effort in rates with two disease models of differing susceptibility to spontaneous hypertension revealed that the IGH region exhibited the highest sequence divergence genome-wide between these lines,58 consistent with the results observed here. Whether increased genetic diversity in human IGHV is associated with a dominant role of the heavy chain in antibody function remains to be seen.
In addition, due to limited genomic resources and accurate high-throughput genotyping tools, there is still much to be learned about the contribution of IG genetic polymorphism to variability in expressed repertoires and the implications of this variation for susceptibility to infectious and autoimmune diseases, responses to therapeutic antibodies and vaccines, and other clinical outcomes. These outstanding questions continue to stress the importance of accurately representing standing genetic variation in the human IG loci.
Materials and methods
Sequencing of IGL and IGK loci from the CHORI-17 BAC library
BAC-end reads from the CHORI-17 hydatidiform mole BAC library mapping to the GRCh37 reference genome were used to identify and select clones within the IGK and IGL loci. The IGL and IGK genes are located at distinct loci in the genome, 22q11.2 and 2p11.2, respectively.1,12,15,23,59,60 Nine clones mapping to the IGL locus spanning chr22: 22209501–23456451 (GRCh37), and eight clones mapping to IGK spanning chr2: 89027491–89630436 (GRCh37), were picked for complete sequencing. As described in Watson et al.6 clones were shotgun sequenced using high quality capillary-based Sanger sequencing and assemblies were constructed and finished on a per clone basis. Fully assembled overlapping BAC clones (Table 1) were then used to create contiguous assemblies spanning the IGK and IGL loci using SeqMan Pro (DNA Star, Lasergene, WI, USA).
Annotation of V, J and C genes and regulatory regions from BAC clones
Sequences of all functional and ORF V, J and C genes (based on IMGT classification) were downloaded from IMGT and Vega databases (www.imgt.org, http://vega.sanger.ac.uk). All sequences were aligned to the completed contigs of each locus using SeqMan Pro, the positions of which were confirmed using BLAST.61 Sequences corresponding to each of the mapped V, J and C genes were extracted from the CH17 contigs, and alleles at each locus were assigned using IMGT V-QUEST.62,63 ‘Novel’ alleles were defined as those not found in the the IMGT reference directory of IMGT/V-QUEST (that contains all functional (F), ORF and in-frame pseudogene (P) alleles from IMGT/GENE-DB).64 To search for potential variants in previously characterized RS and regulatory sequences, SNPs determined from alignments of the CH17 haplotype and assemblies generated previously by Kawasaki et al.15,23 (NG_000834.1, IGK proximal; NG_000833.1. IGK distal; NG_000002.1, IGL) were tested for overlap with 250 bp regions immediately upstream and downstream of functional and ORF gene exons. Since the original assembly of IGK conducted by Kawasaki et al.,23 several additional clones from the RPCI-11 BAC library and a single clone from CH17 have been integrated into the GRCh37 assembly of the distal V gene cluster. In this study, however, to maintain consistency between assembly sequence sources, only clones reported in Kawasaki et al. 23 for the IGK distal region were considered for comparison. Exon coordinates from GRCh37/hg19 as determined by Vega annotations for each gene were downloaded from UCSC (www.genome.ucsc.edu; see Supplementary Tables 1 and 2), and SNP/gene region overlap was assessed using BEDTools version 2.1.65 For those genes in which an SNP was found to occur within the defined regions, sequences in question from the CH17 and reference assemblies (NG_000834.1, IGK proximal; NG_000833.1. IGK distal; NG_000002.1, IGL) were aligned, visually inspected, and compared with previously identified motifs.1
Analyses of structural variants identified in CH17 IGL and IGK BAC clones
Using the program Miropeats66 CH17 contigs for IGK and IGL loci were compared individually to the sequences from assemblies reported by Kawasaki et al.15,23 (IGL, accession NG_000002.1; IGK-proximal/distal, accession NG_000834.1/NG_000833.1); again, only sequence from the original Kawasaki et al.23 assembly were used for comparisons in the distal region of IGK, excluding sequence derived from RPCI-11/CH17 BAC clones. The outputs for each comparison were visually inspected for potential regions of structural variation. Putative breakpoints for the single variant identified were determined by creating a multi-sequence alignment using sequences from the IGL assembly NG_0000002.1 and novel BAC clone that spanned the regions of the predicted variant-associated breakpoints. Multi-sequence alignments were generated and visualized in SeqMan Pro. Gene prediction was carried out using Genscan (genes.mit.edu/GENSCAN.html),67 following by BLAST using the non-redundant gene database.
Comparisons of IGK proximal and distal sequence similarity and recombination analysis
For comparisons of proximal and distal V regions of the IGK locus from both the CH17 and IGK proximal cluster (NG_000834.1) and distal cluster (NG_000833.1) assemblies, only paralogous sequence shared between the proximal and distal regions were considered (that is, sequence spanning the genes IGKV1-6, IGKV1-5, IGKV5-2, and IGKV4-1 was excluded, as these genes do not have paralogous duplicates in the distal IGKV region). Base pair differences were collated based on pair-wise global alignments made between the NG_000834.1 proximal sequence and NG_000833.1 distal sequence from Kawasaki et al.,23 as well as proximal and distal sequences from the CH17 haplotype. Global alignments and variant calls were carried out using ‘run-mummer3’ and ‘combineMUMs’ commands in MUMmer3.0.68 A sequence similarity plot was then generated for each pair-wise comparison using 10 kb windows with a sliding size of 500 bp, as reported previously.23 Sequences, ~22.5 kb in length, from the region suspected to harbor a potential recombination event between proximal and distal regions, were extracted from each haplotype (proximal and distal) and aligned using ClustalW69 within ebioX (http://www.ebioinformatics.org/). Recombination/gene conversion analysis based on this alignment was conducted using the DSS method within TOPALi v2.70,71 This method is based on comparing the branching patterns of two trees constructed using the first and second halves of sequence alignments within a given window of a larger alignment being analyzed; the fit between these two trees and the calculation of DSS is measured using the sum of squares. Windows in which trees differ significantly between the two halves are scored with high DSS values, and are thus candidate sites for recombination. The parameters used here for this analysis were as follows: a window size of 2.5 kb with a step size of 100 bp; the Jukes-Cantor substitution model for calculating distance matrices; 500 bootstrap iterations to test for significance; and the analysis was conducted in both forward and reverse directions along the alignment.
Analysis of IG loci genomic features, locus-wide alignments and SNP discovery
Locus-wide SNPs were first called in the IGLV, IGKV and IGHV regions of the CH17 haplotypes by conducting global alignments of the CH17 and assemblies generated by Kawasaki et al.15,23 and Matsuda et al.38 (NG_000834.1, IGK proximal; NG_000833.1. IGK distal; NG_000002.1, IGL; NG_001019.5 IGH). For IGHV, sequence 10 kb downstream of IGHV6-1 (the most proximal IGHV gene) to 49 kb upstream of IGHV3-74 (the most distal IGHV gene) from both CH17 and NG_001019.56,38 were compared; structural variants identified between the two haplotypes were removed, leaving 834 802 bp of aligned sequence. For IGKV and IGLV, 10 kb downstream of the most proximal V gene and 10 kb upstream of the most distal V gene were compared totalling 858 805 and 858 244 bp of aligned sequence, respectively (the 11.9-kb insertion variant identified in CH17 within IGLV was not included). The lengths of aligned sequence are based on bp coordinates in the reference assemblies (NG_000834.1, IGK proximal; NG_000833.1. IGK distal; NG_000002.1, IGL; NG_001019.5 IGH). CH17 and reference assemblies were aligned on a per locus basis and SNPs were determined from the resulting alignments using the same commands from MUMmer3.0 referenced above. Single-nucleotide variants called from CHM1 cell line DNA using whole-genome paired-end Illumina short-read sequencing39 (NCBI BioProject ID: 176729) were used to assess the accuracy of variants called from the CH17 assemblies. To do this, the NCBI remap tool (http://www.ncbi.nlm.nih.gov/genome/tools/remap) was used to convert the GRCh37 coordinates to the CHM1_1.1 assembly coordinates (GenBank Assembly: GCF_000306695.2), which were then compared with variant calls generated from the alignment of CHM1 Illumina data to the CHM1_1.1 assemblies. Variants in both callsets were flagged as unsupported by the Illumina data, and deemed errors in the CHM1_1.1 assembly.
The coordinates of V gene exon boundaries based on the Vega gene annotation track, repeat content (RepeatMasker 3.2.7; www.repeatmasker.org), SDs,72,73 and centromere/telomere coordinates were downloaded from UCSC (www.genome.ucsc.edu). Percent sequence identity values for SDs were also downloaded from UCSC and used for partitioning the SD data sets. Locus coverage and SNP density values were calculated using BEDTools version 2.1.65 We assessed the statistical significance for the observed enrichments of CH17 SNP densities within SDs in the IGHV and IGKV loci using the Genomic Hyperbrowser (https://hyperbrowser.uio.no40). We created ‘case-control’ tracks including coordinates overlapped (SD) and not overlapped (non-SD) by SDs in the IGHV/IGKV regions, and then carried out tests for enrichments of CH17 SNPs in these loci by permuting the SD and non-SD status for each IGHV and IGKV set of coordinates (MCFDR simulations=10 000). The observed enrichments were then compared with the simulated datasets to calculate P-values for IGHV and IGKV locus analyses. For genome-wide centromere/telomere analysis, we used CHM1 variants,39 and SNP densities in telomeric/centromeric regions were estimated twice independently using telomere/centromere coordinates extended by either 1 or 3 Mb.
GenBank Accessions
CH17-118P18 AC244205.3, CH17-436P12AC243970.3, CH17-405H5 AC245015.2, CH17-140P2 AC244255.3, CH17-158B1 AC233264.2, CH17-98L16 AC245506.3, CH17-216K2 AC243981.3, CH17-68J4 AC247037.2, CH17-335O14 AC245452.3, CH17-424L4 AC245517.3, CH17-264L24 AC245060.1, CH17-238D3 AC245291.4, CH17-242N13 AC246793.1, CH17-420I24 AC244250.3, CH17-475O7 AC244157.2, CH17-95F2 AC245028.2, CH17-355P2 AC245054.3, CHM1_1.1 Assembly GCA_000306695.2.
GenBank Accessions for novel IG alleles
IGLV6-57*02—KM455556, IGLV11-55*02—KM455555, IGLV5-48*02—KM455554, IGLV5-45*04—KM455553, IGLC7*03—KM455557, IGKV1-6*02—KM455558, IGKV1-17*03—KM455566, IGKV6-21*02—KM455568, IGKV2D-26*03—KM455565, IGKV6D-21*02—KM455569, IGKV3D-15*03—KM455564, IGKV1D-13*02—KM455562, IGKV1D-42*02—KM455560, IGKV1D-8*02—KM455563, IGKV1D-8*03—KM455567.
Accession codes
References
Lefranc MP, Lefranc G . The Immunoglobulin Facts Book. Academic Press: London, 2001.
Tonegawa S . Somatic generation of antibody diversity. Nature 1983; 302: 575–581.
Boyd SD, Gaeta BA, Jackson KJ, Fire AZ, Marshall EL, Merker JD et al. Individual variation in the germline Ig gene repertoire inferred from variable region gene rearrangements. J Immunol 2010; 184: 6986–6992.
Glanville J, Kuo TC, von Budingen HC, Guey L, Berka J, Sundar PD et al. Naive antibody gene-segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proc Natl Acad Sci USA 2011; 108: 20066–20071.
Kidd MJ, Chen Z, Wang Y, Jackson KJ, Zhang L, Boyd SD et al. The inference of phased haplotypes for the immunoglobulin H chain V region gene loci by analysis of VDJ gene rearrangements. J Immunol 2012; 188: 1333–1340.
Watson CT, Steinberg KM, Huddleston J, Warren RL, Malig M, Schein J et al. Complete haplotype sequence of the human immunoglobulin heavy-chain variable, diversity, and joining genes and characterization of allelic and copy-number variation. Am J Hum Genet 2013; 92: 530–546.
Watson CT, Breden F . The immunoglobulin heavy chain locus: genetic variation, missing data, and implications for human disease. Genes Immun 2012; 13: 363–373.
Lefranc MP . Immunoglobulin and T cell receptor genes: IMGT((R)) and the birth and rise of immunoinformatics. Front Immunol 2014; 5: 22.
Feeney AJ, Atkinson MJ, Cowan MJ, Escuro G, Lugo G . A defective Vkappa A2 allele in Navajos which may play a role in increased susceptibility to haemophilus influenzae type b disease. J Clin Invest 1996; 97: 2277–2282.
Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 2009; 16: 265–273.
Tsai F-J, Lee Y-C, Chang J-S, Huang L-M, Huang F-Y, Chiu N-C et al. Identification of novel susceptibility Loci for kawasaki disease in a Han chinese population by a genome-wide association study. PLoS ONE 2011; 6: e16853.
Frippiat JP, Williams SC, Tomlinson IM, Cook GP, Cherif D, Le Paslier D et al. Organization of the human immunoglobulin lambda light-chain locus on chromosome 22q11.2. Hum Mol Genet 1995; 4: 983–991.
Kawasaki K, Minoshima S, Schooler K, Kudoh J, Asakawa S, de Jong PJ et al. The organization of the human immunoglobulin lambda gene locus. Genome Res 1995; 5: 125–135.
Williams SC, Frippiat JP, Tomlinson IM, Ignatovich O, Lefranc MP, Winter G . Sequence and evolution of the human germline V lambda repertoire. J Mol Biol 1996; 264: 220–232.
Kawasaki K, Minoshima S, Nakato E, Shibuya K, Shintani A, Schmeits JL et al. One-megabase sequence analysis of the human immunoglobulin lambda gene locus. Genome Res. 1997; 7: 250–261.
Ruiz M, Pallares N, Contet V, Barbi V, Lefranc MP . The human immunoglobulin heavy diversity (IGHD) and joining (IGHJ) segments. Exp Clin Immunogenet 1999; 16: 173–184.
Ghanem N, Dariavach P, Bensmana M, Chibani J, Lefranc G, Lefranc MP . Polymorphism of immunoglobulin lambda constant region genes in populations from France, Lebanon and Tunisia. Exp Clin Immunogenet 1988; 5: 186–195.
Lefranc MP, Pallares N, Frippiat JP . Allelic polymorphisms and RFLP in the human immunoglobulin lambda light chain locus. Hum Genet 1999; 104: 361–369.
Taub RA, Hollis GF, Hieter PA, Korsmeyer S, Waldmann TA, Leder P . Variable amplification of immunoglobulin lambda light-chain genes in human populations. Nature 1983; 304: 172–174.
Cox JP, Tomlinson IM, Winter G . A directory of human germ-line V kappa segments reveals a strong bias in their usage. Eur J Immunol 1994; 24: 827–836.
Huber C, Huber E, Lautner-Rieske A, Schable KF, Zachau HG . The human immunoglobulin kappa locus. Characterization of the partially duplicated L regions. Eur J Immunol 1993; 23: 2860–2867.
Huber C, Schable KF, Huber E, Klein R, Meindl A, Thiebe R et al. The V kappa genes of the L regions and the repertoire of V kappa gene sequences in the human germ line. Eur J Immunol 1993; 23: 2868–2875.
Kawasaki K, Minoshima S, Nakato E, Shibuya K, Shintani A, Asakawa S et al. Evolutionary dynamics of the human immunoglobulin kappa locus and the germline repertoire of the Vkappa genes. Eur J Immunol 2001; 31: 1017–1028.
Osoegawa K, Mammoser AG, Wu C, Frengen E, Zeng C, Catanese JJ et al. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res 2001; 11: 483–496.
Juul L, Hougs L, Barington T . A new apparently functional IGVK gene (VkLa) present in some individuals only. Immunogenetics 1998; 48: 40–46.
Kay PH, Moriuchi J, Ma PJ, Saueracker E . An unusual allelic form of the immunoglobulin lambda constant region genes in the Japanese. Immunogenetics 1992; 35: 341–343.
Moraes Junta C, Passos GA . Genomic EcoRI polymorphism and cosmid sequencing reveal an insertion/deletion and a new IGLV5 allele in the human immunoglobulin lambda variable locus (22q11.2/IGLV). Immunogenetics 2003; 55: 10–15.
Frippiat JP, Dard P, Marsh S, Winter G, Lefranc MP . Immunoglobulin lambda light chain orphons on human chromosome 8q11.2. Eur J Immunol 1997; 27: 1260–1265.
Romo-Gonzalez T, Vargas-Madrazo E . Substitution patterns in alleles of immunoglobulin V genes in humans and mice. Mol Immunol 2006; 43: 731–744.
Pallares N, Lefebvre S, Contet V, Matsuda F, Lefranc MP . The human immunoglobulin heavy variable genes. Exp Clin Immunogenet 1999; 16: 36–60.
Genomes Project Consortium, Genomes Project Consortium Abecasis GR, Genomes Project Consortium Auton A, Genomes Project Consortium Brooks LD, Genomes Project Consortium DePristo MA, Genomes Project Consortium Durbin RM et al. An integrated map of genetic variation from 1092 human genomes. Nature 2012; 491: 56–65.
Brensing-Kuppers J, Zocher I, Thiebe R, Zachau HG . The human immunoglobulin kappa locus on yeast artificial chromosomes (YACs). Gene 1997; 191: 173–181.
Jaenichen HR, Pech M, Lindenmaier W, Wildgruber N, Zachau HG . Composite human VK genes and a model of their evolution. Nucleic Acids Res 1984; 12: 5249–5263.
Barbie V, Lefranc MP . The human immunoglobulin kappa variable (IGKV) genes and joining (IGKJ) segments. Exp Clin Immunogenet 1998; 15: 171–183.
Giudicelli V, Chaume D, Lefranc MP . IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res 2005; 33: D256–D261.
Bailey JA, Eichler EE . Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet 2006; 7: 552–564.
Eichler EE . Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet 2001; 17: 661–669.
Matsuda F, Ishii K, Bourvagnet P, Kuma K, Hayashida H, Miyata T et al. The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J Exp Med 1998; 188: 2151–2162.
Steinberg KM, Schneider VK, Graves-Lindsay TA, Fulton RS, Agarwala R, Huddleston J et al. Single haplotype assembly of the human genome from a hydatidiform mole. Genome Res (in press).
Sandve GK, Gundersen S, Rydbeck H, Glad IK, Holden L, Holden M et al. The Genomic HyperBrowser: inferential genomics at the sequence level. Genome Biol 2010; 11: R121.
Arndt PF, Hwa T, Petrov DA . Substantial regional variation in substitution rates in the human genome: importance of GC content, gene density, and telomere-specific effects. J Mol Evol 2005; 60: 748–763.
Collins AM, Wang Y, Singh V, Yu P, Jackson KJ, Sewell WA . The reported germline repertoire of human immunoglobulin kappa chain genes is relatively complete and accurate. Immunogenetics 2008; 60: 669–676.
Jackson KJL, Wang Y, Gaeta BA, Pomat W, Siba P, Rimmer J et al. Divergent human populations show extensive shared IGK rearrangements in peripheral blood B cells. Immunogenetics 2012; 64: 3–14.
Wang Y, Jackson KJ, Gaeta B, Pomat W, Siba P, Sewell WA et al. Genomic screening by 454 pyrosequencing identifies a new human IGHV gene and sixteen other new IGHV allelic variants. Immunogenetics 2011; 63: 259–265.
Eichler EE . Masquerading repeats: paralogous pitfalls of the human genome. Genome Res 1998; 8: 758–762.
Estivill X, Cheung J, Pujana MA, Nakabayashi K, Scherer SW, Tsui L-C . Chromosomal regions containing high-density and ambiguously mapped putative single nucleotide polymorphisms (SNPs) correlate with segmental duplications in the human genome. Hum Mol Genet 2002; 11: 1987–1995.
Nadel B, Tang A, Lugo G, Love V, Escuro G, Feeney AJ . Decreased frequency of rearrangement due to the synergistic effect of nucleotide changes in the heptamer and nonamer of the recombination signal sequence of the V kappa gene A2b, which is associated with increased susceptibility of Navajos to Haemophilus influenzae type b disease. J Immunol 1998; 161: 6068–6073.
Romo-Gonzalez T, Vargas-Madrazo E . Structural analysis of substitution patterns in alleles of human immunoglobulin VH genes. Mol Immunol 2005; 42: 1085–1097.
Pargent W, Schable KF, Zachau HG . Polymorphisms and haplotypes in the human immunoglobulin kappa locus. Eur J Immunol 1991; 21: 1829–1835.
Schaible G, Rappold GA, Pargent W, Zachau HG . The immunoglobulin kappa locus: polymorphism and haplotypes of Caucasoid and non-Caucasoid individuals. Hum Genet 1993; 91: 261–267.
Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU et al. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet 2005; 77: 78–88.
Hurles ME . Gene conversion homogenizes the CMT1A paralogous repeats. BMC Genomics 2001; 2: 11.
Hallast P, Nagirnaja L, Margus T, Laan M . Segmental duplications and gene conversion: Human luteinizing hormone/chorionic gonadotropin beta gene cluster. Genome Res 2005; 15: 1535–1546.
Verrelli BC, Tishkoff SA . Signatures of selection and gene conversion associated with human color vision variation. Am J Hum Genet 2004; 75: 363–375.
Xu JL, Davis MM . Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity 2000; 13: 37–45.
Avnir Y, Tallarico AS, Zhu Q, Bennett AS, Connelly G, Sheehan J et al. Molecular signatures of hemagglutinin stem-directed heterosubtypic human neutralizing antibodies against influenza A viruses. PLoS Pathog 2014; 10: e1004103.
Tang X-C, Agnihothram SS, Jiao Y, Stanhope J, Graham RL, Peterson EC et al. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc Natl Acad Sci USA 2014; 111: 2018–2026.
Gonzalez-Garay ML, Cranford SM, Braun MC, Doris PA . Diversity in the preimmune immunoglobulin repertoire of SHR lines susceptible and resistant to end-organ injury. Genes Immun (e-pub ahead of print 24 July 2014, doi:10.1038/gene.2014.40).
Zachau HG . The immunoglobulin kappa locus-or-what has been learned from looking closely at one-tenth of a percent of the human genome. Gene 1993; 135: 167–173.
Zachau HG . The immunoglobulin kappa genes. Immunologist 1996; 4: 49–54.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . Basic local alignment search tool. J Mol Biol 1990; 215: 403–410.
Brochet X, Lefranc MP, Giudicelli V . IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res 2008; 36: W503–W508.
Giudicelli V, Brochet X, Lefranc M-P . IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb Protoc 2011; 2011: 695–715.
Alamyar E, Duroux P, Lefranc MP, Giudicelli V . IMGT((R)) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol Biol 2012; 882: 569–604.
Quinlan AR, Hall IM . BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010; 26: 841–842.
Parsons JD . Miropeats: graphical DNA sequence comparisons. Comput Appl Biosci 1995; 11: 615–619.
Burge C, Karlin S . Prediction of complete gene structures in human genomic DNA. J Mol Biol 1997; 268: 78–94.
Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C et al. Versatile and open software for comparing large genomes. Genome Biol 2004; 5: R12.
Thompson JD, Gibson TJ, Higgins DG . Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics 2002; Chapter 2: Unit 2.3.
McGuire G, Wright F . TOPAL 2.0: improved detection of mosaic sequences within multiple alignments. Bioinformatics 2000; 16: 130–134.
Milne I, Wright F, Rowe G, Marshall DF, Husmeier D, McGuire G . TOPALi: software for automatic identification of recombinant sequences within DNA multiple alignments. Bioinformatics 2004; 20: 1806–1807.
Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE . Segmental duplications: organization and impact within the current human genome project assembly. Genome Res 2001; 11: 1005–1017.
Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S et al. Recent segmental duplications in the human genome. Science 2002; 297: 1003–1007.
Acknowledgements
We are grateful to Marie-Paule Lefranc and to the IMGT Nomenclature Committee for their help in defining IG genes and alleles. CTW. was supported in part by a President’s Research Stipend and graduate fellowship awarded by Simon Fraser University. KMS was supported by a Ruth L Kirschstein National Research Service Award (NRSA) training grant to the University of Washington (T32HG00035) and an individual NRSA Fellowship (F32GM097807). This work was supported by the US National Institutes of Health (grants 2R01HG002385 and 5P01HG004120 to EEE) and a National Science and Engineering Research Council of Canada grant to FB. EEE is an Investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
EEE is on the scientific advisory board (SAB) for DNAnexus and was an SAB member of Pacific Biosciences, Inc. (2009-2013) and SynapDx Corp. (2011–2013). The remaining authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Genes and Immunity website
Rights and permissions
About this article
Cite this article
Watson, C., Steinberg, K., Graves, T. et al. Sequencing of the human IG light chain loci from a hydatidiform mole BAC library reveals locus-specific signatures of genetic diversity. Genes Immun 16, 24–34 (2015). https://doi.org/10.1038/gene.2014.56
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2014.56
This article is cited by
-
Characterization of the immunoglobulin lambda chain locus from diverse populations reveals extensive genetic variation
Genes & Immunity (2022)
-
Inter- and intraspecies comparison of phylogenetic fingerprints and sequence diversity of immunoglobulin variable genes
Immunogenetics (2020)
-
On being the right size: antibody repertoire formation in the mouse and human
Immunogenetics (2018)