Abstract
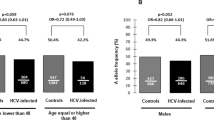
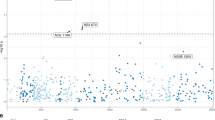
Several data suggest that low-density lipoprotein receptor (LDLR) is a co-receptor for hepatitis C virus (HCV). Soluble LDLR can inhibit HCV infectivity; greater plasma low-density lipoprotein levels are associated with treatment success; LDLR genotypes have a synergistic impact on the likelihood of achieving SVR with Peg-IFN plus RBV, as well as on viral kinetics after starting treatment. The objective of this study was to assess the impact of genetic polymorphisms in genes related to cholesterol synthesis and transport pathways on pre-treatment plasma HCV viral load (VL). A total of 442 patients infected with HCV and treatment naive were prospectively recruited. One hundred forty-four SNPs located in 40 genes from the cholesterol synthesis/transport and IL28B were genotyped and analyzed for genetic association with pre-treatment plasma HCV VL. SNPs rs1433099 and rs2569540 of LDLR showed association with plasma HCV VL (P=4 × 10−4 and P=2 × 10−3) in patients infected with genotypes 1 and 4. A haplotype including the last three exons of LDLR showed association with the cutoff level of 600 000 IU ml−1 VL for genotypes 1 and 4 (OR=0.27; P=8 × 10−6), as well as a quantitative VL (mean±s.d.: 6.19±0.9 vs CC+CG 5.58±1.1 logIU ml−1, P=8 × 10−5). LDLR genotypes are a major genetic factor influencing HCV VL in patients infected with genotypes 1 and 4.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Jones DM, McLauchlan J . Hepatitis C virus: assembly and release of virus particles. J Biol Chem 2010; 285: 22733–22739.
Georgel P, Schuster C, Zeisel MB, Stoll-Keller F, Berg T, Bahram S et al. Virus-host interactions in hepatitis C virus infection: implications for molecular pathogenesis and antiviral strategies. Trends Mol Med 2010; 16: 277–286.
Negro F . Abnormalities of lipid metabolism in hepatitis C virus infection. Gut 2010; 59: 1279–1287.
Woodhouse SD, Narayan R, Latham S, Lee S, Antrobus R, Gangadharan B et al. Transcriptome sequencing, microarray, and proteomic analyses reveal cellular and metabolic impact of hepatitis C virus infection in vitro. Hepatology 2010; 52: 443–453.
Ikeda M, Abe K, Yamada M, Dansako H, Naka K, Kato N . Different anti-HCV profiles of statins and their potential for combination therapy with interferon. Hepatology 2006; 44: 117–125.
Bartenschlager R, Penin F, Lohmann V, Andre P . Assembly of infectious hepatitis C virus particles. Trends Microbiol 2011; 19: 95–103.
Burlone ME, Budkowska A . Hepatitis C virus cell entry: role of lipoproteins and cellular receptors. J Gen Virol 2009; 90: 1055–1070.
Agnello V, Abel G, Elfahal M, Knight GB, Zhang QX . Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci USA 1999; 96: 12766–12771.
Gopal K, Johnson TC, Gopal S, Walfish A, Bang CT, Suwandhi P et al. Correlation between beta-lipoprotein levels and outcome of hepatitis C treatment. Hepatology 2006; 44: 335–340.
Molina S, Castet V, Fournier-Wirth C, Pichard-Garcia L, Avner R, Harats D et al. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J Hepatol 2007; 46: 411–419.
Pineda JA, Caruz A, Di Lello FA, Camacho A, Mesa P, Neukam K et al. Low-density lipoprotein receptor genotyping enhances the predictive value of IL28B genotype in HIV/hepatitis C virus-coinfected patients. AIDS 2011; 25: 1415–1420.
Neukam K, Caruz A, Rivero-Juarez A, Barreiro P, Merino D, Real LM et al. Variations at multiple genes improve interleukin 28b genotype predictive capacity for response to therapy against hepatitis c genotype 1 or 4 infection. AIDS (e-pub ahead of print 9 July 2013; doi:10.1097/01.aids.0000432459.36970.a9).
Rivero-Juarez A, Camacho A, Caruz A, Neukam K, Gonzalez R, Di Lello FA et al. LDLr genotype modifies the impact of IL28B on HCV viral kinetics after the first weeks of treatment with PEG-IFN/RBV in HIV/HCV patients. AIDS 2012; 26: 1009–1015.
Albecka A, Belouzard S, Op de Beeck A, Descamps V, Goueslain L, Bertrand-Michel J et al. Role of low-density lipoprotein receptor in the hepatitis C virus life cycle. Hepatology; 55: 998–1007.
Mackenzie JM, Khromykh AA, Parton RG . Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe 2007; 2: 229–239.
Mas Marques A, Mueller T, Welke J, Taube S, Sarrazin C, Wiese M et al. Low-density lipoprotein receptor variants are associated with spontaneous and treatment-induced recovery from hepatitis C virus infection. Infect Genet Evol 2009; 9: 847–852.
Hennig BJ, Hellier S, Frodsham AJ, Zhang L, Klenerman P, Knapp S et al. Association of low-density lipoprotein receptor polymorphisms and outcome of hepatitis C infection. Genes Immun 2002; 3: 359–367.
Haqqani AA, Tilton JC . Entry inhibitors and their use in the treatment of HIV-1 infection. Antiviral Res 2013; 98: 158–170.
Sainz B Jr, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S et al. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med; 18: 281–285.
Mosbruger TL, Duggal P, Goedert JJ, Kirk GD, Hoots WK, Tobler LH et al. Large-scale candidate gene analysis of spontaneous clearance of hepatitis C virus. J Infect Dis 2010; 201: 1371–1380.
Agundez JA, Garcia-Martin E, Devesa MJ, Carballo M, Martinez C, Lee-Brunner A et al. Polymorphism of the TLR4 gene reduces the risk of hepatitis C virus-induced hepatocellular carcinoma. Oncology 2012; 82: 35–40.
Danilovic DL, Mendes-Correa MC, Lima EU, Zambrini H, R KB, Marui S . Correlations of CTLA-4 gene polymorphisms and hepatitis C chronic infection. Liver Int 2012; 32: 803–808.
Li H, Liu Z, Han Q, Li Y, Chen J . Association of genetic polymorphism of low-density lipoprotein receptor with chronic viral hepatitis C infection in Han Chinese. J Med Virol 2006; 78: 1289–1295.
Anand SS, Xie C, Pare G, Montpetit A, Rangarajan S, McQueen MJ et al. Genetic variants associated with myocardial infarction risk factors in over 8000 individuals from five ethnic groups: the INTERHEART genetics study. Circ Cardiovasc Genet 2009; 2: 16–25.
Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010; 466: 707–713.
Muallem H, North KE, Kakoki M, Wojczynski MK, Li X, Grove M et al. Quantitative effects of common genetic variations in the 3′UTR of the human LDL-receptor gene and their associations with plasma lipid levels in the Atherosclerosis Risk in Communities study. Hum Genet 2007; 121: 421–431.
Park CY, Jun HJ, Wakita T, Cheong JH, Hwang SB . Hepatitis C virus nonstructural 4B protein modulates sterol regulatory element-binding protein signaling via the AKT pathway. J Biol Chem 2009; 284: 9237–9246.
Waris G, Felmlee DJ, Negro F, Siddiqui A . Hepatitis C virus induces proteolytic cleavage of sterol regulatory element binding proteins and stimulates their phosphorylation via oxidative stress. J Virol 2007; 81: 8122–8130.
Rivero-Juarez A, Camacho Espejo A, Perez-Camacho I, Neukam K, Caruz A, Mira JA et al. Association between the IL28B genotype and hepatitis C viral kinetics in the early days of treatment with pegylated interferon plus ribavirin in HIV/HCV co-infected patients with genotype 1 or 4. J Antimicrob Chemother 2012; 67: 202–205.
Schectman G, Kaul S, Mueller RA, Borden EC, Kissebah AH . The effect of interferon on the metabolism of LDLs. Arterioscler Thromb 1992; 12: 1053–1062.
Fischer DG, Novick D, Cohen B, Rubinstein M . Isolation and characterization of a soluble form of the LDL receptor, an interferon-induced antiviral protein. Proc Soc Exp Biol Med 1994; 206: 228–232.
Fischer DG, Tal N, Novick D, Barak S, Rubinstein M . An antiviral soluble form of the LDL receptor induced by interferon. Science 1993; 262: 250–253.
Albecka A, Belouzard S, Op de Beeck A, Descamps V, Goueslain L, Bertrand-Michel J et al. Role of low-density lipoprotein receptor in the hepatitis C virus life cycle. Hepatology 2012; 55: 998–1007.
Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. Embo J 2002; 21: 5017–5025.
Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet; 45: 164–171.
Nyholt DR . A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet 2004; 74: 765–769.
Sasieni PD . From genotypes to genes: doubling the sample size. Biometrics 1997; 53: 1253–1261.
Acknowledgements
This study has been supported in part by grants from the Spanish Health Ministry (ISCIII-RETIC RD06/006 and projects PI10/01664 and PI10/01232) from the Consejería de Salud de la Junta de Andalucía (PI-0247-2010) and from the Fundación para la Investigación y la Prevención del Sida en España (reference 360799/09). JAP is the recipient of an extension grant from the Instituto de Salud Carlos III (grant number Programa-I3SNS). KN is the recipient of a ‘Sara Borrell’ postdoctoral perfection grant from the Instituto de Salud Carlos III (SCO/523/2008).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Genes and Immunity website
Supplementary information
Rights and permissions
About this article
Cite this article
Caruz, A., Neukam, K., Rivero-Juárez, A. et al. Association of low-density lipoprotein receptor genotypes with hepatitis C viral load. Genes Immun 15, 16–24 (2014). https://doi.org/10.1038/gene.2013.56
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2013.56
Keywords
This article is cited by
-
Genetic markers of lipid metabolism genes associated with low susceptibility to HCV infection
Scientific Reports (2019)
-
APOB codon 4311 polymorphism is associated with hepatitis C virus infection through altered lipid metabolism
BMC Gastroenterology (2018)
-
Impact of selenium supplementation on fish antiviral responses: a whole transcriptomic analysis in rainbow trout (Oncorhynchus mykiss) fed supranutritional levels of Sel-Plex®
BMC Genomics (2016)