Abstract
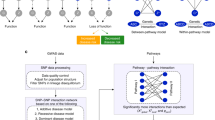
Gene–gene interactions are proposed as an important component of the genetic architecture of complex diseases, and are just beginning to be evaluated in the context of genome-wide association studies (GWAS). In addition to detecting epistasis, a benefit to interaction analysis is that it also increases power to detect weak main effects. We conducted a knowledge-driven interaction analysis of a GWAS of 931 multiple sclerosis (MS) trios to discover gene–gene interactions within established biological contexts. We identify heterogeneous signals, including a gene–gene interaction between CHRM3 (muscarinic cholinergic receptor 3) and MYLK (myosin light-chain kinase) (joint P=0.0002), an interaction between two phospholipase C-β isoforms, PLCβ1 and PLCβ4 (joint P=0.0098), and a modest interaction between ACTN1 (actinin alpha 1) and MYH9 (myosin heavy chain 9) (joint P=0.0326), all localized to calcium-signaled cytoskeletal regulation. Furthermore, we discover a main effect (joint P=5.2E−5) previously unidentified by single-locus analysis within another related gene, SCIN (scinderin), a calcium-binding cytoskeleton regulatory protein. This work illustrates that knowledge-driven interaction analysis of GWAS data is a feasible approach to identify new genetic effects. The results of this study are among the first gene–gene interactions and non-immune susceptibility loci for MS. Further, the implicated genes cluster within inter-related biological mechanisms that suggest a neurodegenerative component to MS.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Barcellos LF, Sawcer S, Ramsay PP, Baranzini SE, Thomson G, Briggs F et al. Heterogeneity at the HLA-DRB1 locus and risk for multiple sclerosis. Hum Mol Genet 2006; 15: 2813–2824.
The International Multiple Sclerosis Genetics Consortium. A second major histocompatibility complex susceptibility locus for multiple sclerosis. Ann Neurol 2007; 61: 228–236.
Cordell HJ . Genome-wide association studies: detecting gene-gene interactions that underlie human diseases. Nat Rev Genet 2009; 10: 392–404.
Marchini J, Donnelly P, Cardon LR . Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet 2005; 37: 413–417.
Rodriguez Del CA, Vitale ML, Tchakarov L, Trifaro JM . Human platelets contain scinderin, a Ca(2+)-dependent actin filament-severing protein. Thromb Haemost 1992; 67: 248–251.
Trifaro JM, Rose SD, Marcu MG . Scinderin, a Ca2+-dependent actin filament severing protein that controls cortical actin network dynamics during secretion. Neurochem Res 2000; 25: 133–144.
Greene CS, Penrod NM, Williams SM, Moore JH . Failure to replicate a genetic association may provide important clues about genetic architecture. PLoS One 2009; 4: e5639.
Fraley TS, Pereira CB, Tran TC, Singleton C, Greenwood JA . Phosphoinositide binding regulates alpha-actinin dynamics: mechanism for modulating cytoskeletal remodeling. J Biol Chem 2005; 280: 15479–15482.
Kremerskothen J, Teber I, Wendholt D, Liedtke T, Bockers TM, Barnekow A . Brain-specific splicing of alpha-actinin 1 (ACTN1) mRNA. Biochem Biophys Res Commun 2002; 295: 678–681.
Divers J, Freedman BI . Susceptibility genes in common complex kidney disease. Curr Opin Nephrol Hypertens 2010; 19: 79–84.
Mhatre AN, Janssens S, Nardi MA, Li Y, Lalwani AK . Clinical and molecular genetic analysis of a family with macrothrombocytopenia and early onset sensorineural hearing loss. Eur J Med Genet 2009; 52: 185–190.
Geguchadze R, Zhi G, Lau KS, Isotani E, Persechini A, Kamm KE et al. Quantitative measurements of Ca(2+)/calmodulin binding and activation of myosin light chain kinase in cells. FEBS Lett 2004; 557: 121–124.
De KJ, Wilczak N, Leta R, Streetland C . Astrocytes in multiple sclerosis lack beta-2 adrenergic receptors. Neurology 1999; 53: 1628–1633.
Nizri E, Hamra-Amitay Y, Sicsic C, Lavon I, Brenner T . Anti-inflammatory properties of cholinergic up-regulation: a new role for acetylcholinesterase inhibitors. Neuropharmacology 2006; 50: 540–547.
Jian X, Szaro BG, Schmidt JT . Myosin light chain kinase: expression in neurons and upregulation during axon regeneration. J Neurobiol 1996; 31: 379–391.
Gallo G . Myosin II activity is required for severing-induced axon retraction in vitro. Exp Neurol 2004; 189: 112–121.
Lukas TJ, Miao H, Chen L, Riordan SM, Li W, Crabb AM et al. Susceptibility to glaucoma: differential comparison of the astrocyte transcriptome from glaucomatous African American and Caucasian American donors. Genome Biol 2008; 9: R111.
Akkad DA, Hoffjan S, Petrasch-Parwez E, Beygo J, Gold R, Epplen JT . Variation in the IL7RA and IL2RA genes in German multiple sclerosis patients. J Autoimmun 2009; 32: 110–115.
Alcina A, Fedetz M, Ndagire D, Fernandez O, Leyva L, Guerrero M et al. IL2RA/CD25 gene polymorphisms: uneven association with multiple sclerosis (MS) and type 1 diabetes (T1D). PLoS One 2009; 4: e4137.
Rubio JP, Stankovich J, Field J, Tubridy N, Marriott M, Chapman C et al. Replication of KIAA0350, IL2RA, RPL5 and CD58 as multiple sclerosis susceptibility genes in Australians. Genes Immun 2008; 9: 624–630.
Mills GB, Cheung RK, Grinstein S, Gelfand EW . Increase in cytosolic free calcium concentration is an intracellular messenger for the production of interleukin 2 but not for expression of the interleukin 2 receptor. J Immunol 1985; 134: 1640–1643.
Suh PG, Park JI, Manzoli L, Cocco L, Peak JC, Katan M et al. Multiple roles of phosphoinositide-specific phospholipase C isozymes. BMB Rep 2008; 41: 415–434.
Adamski FM, Timms KM, Shieh BH . A unique isoform of phospholipase Cbeta4 highly expressed in the cerebellum and eye. Biochim Biophys Acta 1999; 1444: 55–60.
Homma Y, Takenawa T, Emori Y, Sorimachi H, Suzuki K . Tissue- and cell type-specific expression of mRNAs for four types of inositol phospholipid-specific phospholipase C. Biochem Biophys Res Commun 1989; 164: 406–412.
Kim D, Jun KS, Lee SB, Kang NG, Min DS, Kim YH et al. Phospholipase C isozymes selectively couple to specific neurotransmitter receptors. Nature 1997; 389: 290–293.
Ban M, Goris A, Lorentzen AR, Baker A, Mihalova T, Ingram G et al. Replication analysis identifies TYK2 as a multiple sclerosis susceptibility factor. Eur J Hum Genet 2009; 17: 1309–1313.
De Jager PL, Jia X, Wang J, de Bakker PI, Ottoboni L, Aggarwal NT et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet 2009; 41: 776–782.
Gregory SG, Schmidt S, Seth P, Oksenberg JR, Hart J, prokop A et al. Interleukin 7 receptor alpha chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat Genet 2007; 39: 1083–1091.
The International Multiple Sclerosis Genetics Consortium. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med 2007; 357: 851–862.
Baranzini SE, Galwey NW, Wang J, Khankhanian P, Lindberg R, Pelletier D et al. Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Hum Mol Genet 2009; 18: 2078–2090.
Elbers CC, van Eijk KR, Franke L, Mulder F, van der Schouw YT, Wijmenga C et al. Using genome-wide pathway analysis to unravel the etiology of complex diseases. Genet Epidemiol 2009; 33: 419–431.
O’Dushlaine C, Kenny E, Heron EA, Segurado R, Gill M, Morris DW et al. The SNP ratio test: pathway analysis of genome-wide association datasets. Bioinformatics 2009; 25: 2762–2763.
Peng G, Luo L, Siu H, Zhu Y, Hu P, Hong S et al. Gene and pathway-based second-wave analysis of genome-wide association studies. Eur J Hum Genet 2009; 18: 111–117.
Saccone SF, Saccone NL, Swan GE, Madden PA, Goate AM, Rice JP et al. Systematic biological prioritization after a genome-wide association study: an application to nicotine dependence. Bioinformatics 2008; 24: 1805–1811.
Torkamani A, Topol EJ, Schork NJ . Pathway analysis of seven common diseases assessed by genome-wide association. Genomics 2008; 92: 265–272.
Bush WS, Dudek SM, Ritchie MD . Biofilter: a knowledge-integration system for the multi-locus analysis of genome-wide association studies. Pac Symp Biocomput 2009; 14: 368–379.
Cordell HJ, Barratt BJ, Clayton DG . Case/pseudocontrol analysis in genetic association studies: a unified framework for detection of genotype and haplotype associations, gene-gene and gene-environment interactions, and parent-of-origin effects. Genet Epidemiol 2004; 26: 167–185.
Fisher RA . The correlation between relatives on the supposition of mendelian inheritance. Trans R Soc Edinburgh 1918; 52: 399–433.
Acknowledgements
This research was funded by the NIH grants 1R01 LM010040-01 and 5R01 NS049477-05. This study makes use of data generated by the WTCCC.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Genes and Immunity website
Supplementary information
Rights and permissions
About this article
Cite this article
Bush, W., McCauley, J., DeJager, P. et al. A knowledge-driven interaction analysis reveals potential neurodegenerative mechanism of multiple sclerosis susceptibility. Genes Immun 12, 335–340 (2011). https://doi.org/10.1038/gene.2011.3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2011.3
Keywords
This article is cited by
-
Protease-activated receptor-1 activation by granzyme B causes neurotoxicity that is augmented by interleukin-1β
Journal of Neuroinflammation (2017)
-
Identifying gene–gene interactions that are highly associated with four quantitative lipid traits across multiple cohorts
Human Genetics (2017)
-
Genomic analyses with biofilter 2.0: knowledge driven filtering, annotation, and model development
BioData Mining (2013)
-
Gene ontology analysis of pairwise genetic associations in two genome-wide studies of sporadic ALS
BioData Mining (2012)