Abstract
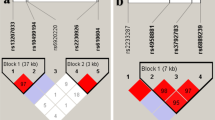
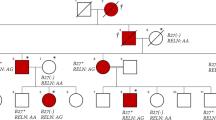
Associations with disease identified by genome-wide association studies (GWAS) must be replicated and refined to validate causative variants. In the Wellcome Trust Case Control Consortium (WTCCC) GWAS using 14 500 non-synonymous single nucleotide polymorphisms (nsSNP), rs11062385 (a nsSNP in JARID1A) showed nominal association with ankylosing spondylitis (AS) (P=0.0006, odds ratio (OR)=1.26, 95% confidence interval (95% CI)=1.1–1.4). To replicate and refine the association of JARID1A, rs11062385 was genotyped in 730 further cases and compared with allele frequencies in non-AS disease cohorts typed by WTCCC. We replicated the initial association (P=0.04, OR=1.16, 95% CI=1.01–1.34) and identified a strengthened association with AS in a meta-analysis of this new study combined with the original WTCCC study (P=0.0001, OR=1.21, 95% CI=1.10–1.33). We also genotyped nine further intronic tagging SNPs in JARID1A in 1604 AS cases and 1020 new control samples, but none was associated with AS. JARID1A or a locus in strong linkage disequilibrium with it is a positional candidate for susceptibility to AS.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Pociot F, Akolkar B, Concannon P, Erlich HA, Julier C, Morahan G et al. Genetics of type 1 diabetes: What's next? Diabetes 2010; 59: 1561–1571.
Gregersen PK . Susceptibility genes for rheumatoid arthritis A rapidly expanding harvest. Bull NYU Hosp Jt Dis 2010; 68: 179–182.
Wellcome Trust Case Control Consortium and The Australo-Anglo-American Spondylitis Consortium. Association scan of 14 500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet 2007; 39: 1329–1337.
Harvey D, Pointon JJ, Evans DM, Karaderi T, Farrar C, Appleton LH et al. Investigating the genetic association between ERAP1 and ankylosing spondylitis. Hum Mol Genet 2009; 18: 4204–4212.
Karaderi T, Harvey D, Farrar C, Appleton LH, Stone MA, Sturrock RD et al. Association between the interleukin 23 receptor and ankylosing spondylitis is confirmed by a new UK case-control study and meta-analysis of published series. Rheumatology (Oxford) 2009; 48: 386–389.
Pointon JJ, Harvey D, Karaderi T, Appleton LH, Farrar C, Stone MA et al. Elucidating the chromosome 9 association with AS; CARD9 is a candidate gene. Genes Immun 2010; 11: 490–496.
Pointon JJ, Harvey D, Karaderi T, Appleton LH, Farrar C, Stone MA et al. The chromosome 16q region associated with ankylosing spondylitis includes the candidate gene tumor necrosis factor receptor type 1-associated death domain (TRADD). Ann Rheum Dis 2010; 69: 1243–1246.
Defeo-Jones D, Huang PS, Jones RE, Haskell KM, Vuocolo GA, Hanobik MG et al. Cloning of cDNAs for cellular proteins that bind to the retinoblastoma gene product. Nature 1991; 352: 251–254.
Chan SW, Hong W . Retinoblastoma-binding protein 2 (Rbp2) potentiates nuclear hormone receptor-mediated transcription. J Biol Chem 2001; 276: 28402–28412.
Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA 2002; 99: 4465–4470.
Van der Linden S, Valkenburg HA, Cats A . Evaluation of diagnostic criteria for ankylosing spondylitis. Arthritis Rheum 1984; 27: 361–368.
Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC et al. A genome-wide association study of global gene expression. Nat Genet 2007; 39: 1202–1207.
Lu Q, Kaplan M, Ray D, Ray D, Zacharek S, Gutsch D et al. Demethylation of ITGAL (CD11a) regulatory sequences in systemic lupus erythematosus. Arthritis Rheum 2002; 46: 1282–1291.
Mi XB, Zeng FQ . Hypomethylation of interleukin-4 and -6 promoters in T cells from systemic lupus erythematosus patients. Acta Pharmacol Sin 2008; 29: 105–112.
Hu N, Qiu X, Luo Y, Yuan J, Li Y, Lei W et al. Abnormal histone modification patterns in Lupus CD4+ T cells. J Rheum 2008; 35: 804–810.
Fuks K, Hurd PJ, Deplus R, Kouzarides T . The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucl Acids Res 2003; 31: 2305–2312.
van Zutven LJ, Onen E, Velthuizen SC, van Drunen E, von Bergh AR, van den Heuvel-Eibrink MM et al. Identification of NUP98 abnormalities in acute leukemia: JARID1A (12p13) as a new partner gene. Genes Chrom Cancer 2006; 45: 437–446.
Richardson B . Effect of an inhibitor of DNA methylation on T cells. II.5-Azacytidine induces self-reactivity in antigen-specific T4+ cells. Hum Immunol 1986; 17: 456–470.
Spannhoff A, Hauser A-T, Heinke R, Sippl W, Jung M . The emerging therapeutic potential of histone methyltransferase and demethylase inhibitors. ChemMedChem 2009; 4: 1568–1582.
Acknowledgements
JP is funded by the NIHR Oxford Biomedical Research Centre ankylosing spondylitis chronic disease cohort (Theme Code: A91202). DH is funded by Arthritis Research UK (Grant no: 19356). CF is funded by the Thames Valley Comprehensive Local Research Network (TVCLRN), which forms part of the NIHR Comprehensive Clinical Research Network (CCRN). TK is funded by the Henni Mester studentship. This study was also funded, in part, by the Arthritis Research UK, award numbers 18797 and by the Wellcome Trust under award no. 076113. We are grateful to the many patients who contributed samples to these studies and to their physicians for allowing us to study their patients. We also thank the National Ankylosing Spondylitis Society (UK) for additional financial support for DH and their unstinting help in patient recruitment. We thank the Osteoarthritis Group, Oxford, for their control samples. This study makes use of data generated by the Wellcome Trust Case Control Consortium. A full list of the investigators who contributed to the data is available at http://www.wtccc.org.uk. We acknowledge the use of genotype data from the British 1958 Birth Cohort DNA collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02. Additional laboratory support was provided by the NIHR Oxford Musculoskeletal Biomedical Research Unit. Finally, we thank Lyn-Louise Johnson for iPLEX genotyping and the Bioinformatics and Statistical Genetics groups for support, both of the Wellcome Trust Centre for Human Genetics, Oxford.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Pointon, J., Harvey, D., Karaderi, T. et al. The histone demethylase JARID1A is associated with susceptibility to ankylosing spondylitis. Genes Immun 12, 395–398 (2011). https://doi.org/10.1038/gene.2011.23
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2011.23
Keywords
This article is cited by
-
Beyond receptors and signaling: epigenetic factors in the regulation of innate immunity
Immunology & Cell Biology (2015)
-
Histone lysine demethylases as targets for anticancer therapy
Nature Reviews Drug Discovery (2013)
-
Molecular mechanisms and potential functions of histone demethylases
Nature Reviews Molecular Cell Biology (2012)