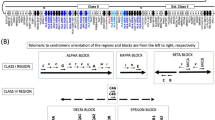
Abstract
The major histocompatibility complex (MHC) (Chromosome 6p21.3) is a dynamic, immune gene-rich region that is associated with multiple diseases. Haplotype-tagging single-nucleotide polymorphism (htSNP) panels for the MHC can aid association studies but have only been reported for African, Asian and Caucasian populations to date. We genotyped 2154 SNPs spanning a 3.8-Mb region of the classical MHC in 94 healthy African Americans using Illumina BeadArray technology. We describe the haplotype structure of the MHC in African Americans, calculate the recombination rate (0.35 cM Mb−1) across the region, identify recombination hot spots and develop a panel of htSNPs for future genetic association studies in this population. We conclude that while patterns of LD and recombination are similar within the MHC to that reported in other populations, differences in minor allele frequency at specific markers necessitates an htSNP panel unique to African Americans, which we provide here for use in future genetic association studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK et al. Gene map of the extended human MHC. Nat Rev Genet 2004; 5: 889–899.
Kelley J, Trowsdale J . Features of MHC and NK gene clusters. Transpl Immunol 2005; 14: 129–134.
de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet 2006; 38: 1166–1172.
Miretti MM, Walsh EC, Ke X, Delgado M, Griffiths M, Hunt S et al. A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms. Am J Hum Genet 2005; 76: 634–646.
Stewart CA, Horton R, Allcock RJ, Ashurst JL, Atrazhev AM, Coggill P et al. Complete MHC haplotype sequencing for common disease gene mapping. Genome Res 2004; 14: 1176–1187.
Stenzel A, Lu T, Koch WA, Hampe J, Guenther SM, De La Vega FM et al. Patterns of linkage disequilibrium in the MHC region on human chromosome 6p. Hum Genet 2004; 114: 377–385.
Gonzalez-Neira A, Ke X, Lao O, Calafell F, Navarro A, Comas D et al. The portability of tagSNPs across populations: a worldwide survey. Genome Res 2006; 16: 323–330.
Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR et al. Methods for high-density admixture mapping of disease genes. Am J Hum Genet 2004; 74: 979–1000.
Tian C, Hinds DA, Shigeta R, Kittles R, Ballinger DG, Seldin MF . A genomewide single-nucleotide-polymorphism panel with high ancestry information for African American admixture mapping. Am J Hum Genet 2006; 79: 640–649.
Hughes LB, Morrison D, Kelley JM, Padilla MA, Vaughn LK, Westfall AO et al. The HLA-DRB1 shared epitope is associated with susceptibility to rheumatoid arthritis and with percentage of European genetic ancestry in African-Americans. Arthritis Rheum 2008; 58: 349–358.
Parham P, Ohta T . Population biology of antigen presentation by MHC class I molecules. Science 1996; 272: 67–74.
Hedrick PW . Pathogen resistance and genetic variation at MHC loci. Evolution Int J Org Evolution 2002; 56: 1902–1908.
Stephens M, Donnelly P . A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 2003; 73: 1162–1169.
McVean G, Awadalla P, Fearnhead P . A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics 2002; 160: 1231–1241.
Acknowledgements
We thank Andrew O Westfall, MS; Graciela S Alarcón, MD, MPH; Stephanie Ledbetter, MS; Cynthia Irwin, RN, MPH; Eugene Oliver, BS; Selena Luckett, RN, CRNC; Laticia Woodruff, RN, MSN; Stephanie McLean, BS; Zenoria Causey (UAB); Doyt L Conn, MD; Joyce Carlone, RN, RNP; Karla Caylor, BSN, RN; Meri Eger, RN (Emory); Beth L Jonas, MD; Leigh F Callahan, PhD; Pat Cummins, RN (UNC); Edwin A Smith, MD; Gary Gilkeson, MD; Trisha Sturgil (MUSC); and Larry W Moreland, MD (University of Pittsburgh) for efforts in participant recruitment. We also appreciate the technical assistance of Jinyi Wang and Yuanqing Edberg. We thank David B Allison, PhD (UAB) and Matthew Stephens, PhD (University of Washington) for advice on statistical methods. This work was supported by NIH/NIAMS N01 AR02247, NIH R01 AR51394 and GCRC Grant M01 RR00032 (University of Alabama at Birmingham) from the National Center for Research Resources. JMK and LKV were supported by NIH T32 AR07450 (NIAMS). MAP was supported in part by the Research Supplements for Underrepresented Minorities Program (R01 AR052658-02S1).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Genes and Immunity website (http://www.nature.com/gene)
Supplementary information
Rights and permissions
About this article
Cite this article
Kelley, J., Hughes, L., Feng, R. et al. Evaluating linkage disequilibrium and recombination provides a haplotype-tagging SNP panel of the major histocompatibility complex in African Americans. Genes Immun 9, 271–273 (2008). https://doi.org/10.1038/gene.2008.6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2008.6