Abstract
Embryonic stem (ES) cells are pluripotent cells characterized by self-renewability and differentiation potential. Induced pluripotent stem (iPS) cells are ES cell-equivalent cells derived from somatic cells by the introduction of core reprogramming factors. ES and iPS cells are important sources for understanding basic biology and for generating therapeutic cells for clinical applications. Tribbles homolog 2 (Trib2) functions as a scaffold in signaling pathways. However, the relevance of Trib2 to the pluripotency of ES and iPS cells is unknown. In the present study, we elucidated the importance of Trib2 in maintaining pluripotency in mouse ES cells and in generating iPS cells from somatic cells through the reprogramming process. Trib2 expression decreased as ES cells differentiated, and Trib2 knockdown in ES cells changed their colony morphology while reducing the activity of alkaline phosphatase and the expression of the pluripotency marker genes Oct4, Sox2, Nanog and Klf4. Trib2 directly interacted with Oct4 and elevated Oct4 promoter activity. During the generation of iPS cells, Trib2 knockdown decreased the reprogramming efficiency of mouse embryonic fibroblasts, whereas Trib2 overexpression significantly increased their reprogramming efficiency. In summary, our results suggest that Trib2 is important for maintaining self-renewal in ES cells and for pluripotency induction during the reprogramming process.
Similar content being viewed by others
Introduction
Pluripotent cells have drawn major attention due to their nearly unlimited self-renewal potential and their ability to differentiate into all types of cells in the body.1, 2 They are broadly classified into two main groups: embryonic stem (ES) cells, which are derived from the inner cell masses (ICMs) of mammalian blastocysts, and induced pluripotent stem (iPS) cells, which can be generated from somatic cells by introducing defined transcription factors, such as Oct4, Sox2, Klf4 and c-Myc.3 iPS cells are molecularly and functionally similar to ES cells.4, 5 ES cells and iPS cells are attractive for therapeutic applications due to their unlimited potential to self-renew and differentiate.6, 7, 8 Therefore, an understanding of the molecular mechanism underlying pluripotency induction and maintenance has a significant impact on basic biology and on clinical applications for treating various diseases using regenerative medicine.
Tribbles homolog 2 (Trib2) is a member of the Tribbles family, and members of this family act as adaptors or scaffolds in signaling pathways.9 Trib2 is located mostly in the cytoplasm of several types of cells.10 In lung cancer, Trib2 interacts with TRIM21, an E3 ubiquitin ligase that degrades C/EBPα, which inhibits tumor growth in vivo. Trib2 knockdown leads to morphological changes in lung cancer cells that resemble C/EBPα overexpression.11 Trib2 is associated with cancer progression and tumorigenesis. Trib2 is highly expressed in malignant melanoma, and silencing Trib2 decreases colony formation and migration ability.12 Although Trib2 is implicated in multiple important cellular processes, its relevance in pluripotency regulation has not been reported.
In the present study, for the first time, we have examined the roles of Trib2 in maintaining the self-renewal of ES cells and inducing somatic cell pluripotency through the reprogramming process. This research demonstrates a novel function of Trib2 in regulating pluripotency and describes the mechanism of Trib2 and Oct4 interaction.
Materials and methods
Materials
Dulbecco's modified Eagle medium (DMEM), non-essential amino acids, Glutamax-1, fetal bovine serum (FBS) trypsin, Lipofectamine-plus, Blasticidin and the Alexa Fluor 488 goat anti-rabbit (A-11034), Alexa Fluor 488 goat anti-mouse (A-11029), Alexa Fluor 488 donkey anti-goat (A-11055), and Alexa Fluor 568 goat anti-rabbit (A-11011) antibodies were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Leukemia inhibitory factor (LIF), the anti-GAPDH antibody (MAB374), and a control antibody for chromatin immunoprecipitation (NI02) were purchased from EMD Millipore Corp. (Billerica, MA, USA). Penicillin-streptomycin was purchased from HyClone (Logan, UT, USA). Culture plates were purchased from Nunc (Roskilde, Denmark). Vectashield mounting medium with 4′-6-Diamidino-2-phenylindole was purchased from Vector Laboratories (Burlingame, CA, USA). Peroxidase-labeled secondary antibodies and enhanced chemiluminescence Western blotting detection reagents were purchased from Amersham Biosciences (Pittsburgh, PA, USA). The pNTAP vector (#240101) was purchased from Stratagene (La Jolla, CA, USA). Streptavidin resin (#240105) was purchased from Agilent Technologies, Inc. (Santa Clara, CA, USA). The following antibodies were used in this study: anti-Sox2 (5ab 9776), purchased from Abcam (Cambridge, MA, USA); anti-Trib2 (15359-1-AP), purchased from Proteintech Group (Manchester, UK); anti-Nanog (#8822), purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA); anti-SSEA-1 (sc-21702), anti-Oct4 (sc-8628), and anti-Sox2 (sc-17320), purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA); and anti-FLAG-tag (F9291), purchased from Sigma-Aldrich (St Louis, MO, USA). The Dual-Luciferase Reporter Assay System and the pGL3-basic vector (E1751) were purchased from Promega (Madison, WI, USA). Protease inhibitor cocktail (P8849), β-mercaptoethanol (M6250), an alkaline phosphatase detection kit (86R) and all other chemicals were purchased from Sigma-Aldrich.
Cell culture and differentiation assays
D3 and OG2/ROSA26 mouse ES (mES) cells were cultured on mitomycin C-treated mouse embryonic fibroblasts (MEFs) in ES cell culture media (DMEM supplemented with 1 × non-essential amino acids, 1 × Glutamax-1, 15% FBS, 1 × penicillin-streptomycin, 0.1 mM β-mercaptoethanol, and 1000 Uml−1 LIF). MEFs were isolated from 12.5-day-old embryos from pregnant C3H mice and maintained in MEF medium (DMEM with high glucose, 1 × Glutamax-1, 10% FBS, and 1 × penicillin-streptomycin) as described previously.3 To culture embryoid bodies (EBs) via the hanging-drop method, 20 μl (50 000 cells per ml) of dissociated ES cells were spotted on a Petri dish lid in ES cell medium without LIF. After 4 days, EBs were transferred to a 1% gelatin-coated dish to induce their spontaneous differentiation and then incubated in ES cell medium without LIF for 6 days. Cells were incubated in a humidified incubator at 37 °C under 5% CO2.
Gene silencing using shRNA lentivirus
pLKO.1-puro lentiviral vectors expressing Trib2 shRNA (TRCN0000362058) and non-target control shRNA (SHC002) were purchased from Sigma-Aldrich. To generate lentiviral particles, 293FT cells were co-transfected with the shRNA lentiviral plasmid (pLKO.1-puro) and ViraPower Lentiviral Packaging Mix (pLP1, pLP2 and pLP-VSV-G; Thermo Fisher Scientific, Waltham, MA, USA) using Lipofectamine-plus and culture supernatants containing lentivirus collected 48 h after transfection. For lentiviral transduction, D3 mES cells were treated with culture supernatants from 293FT cells in the presence of 10 μg ml−1 polybrene (Sigma-Aldrich), and stable cell lines expressing shRNA were generated by selection with puromycin (5 μg ml−1). To ensure shRNA-mediated Trib2 expression silencing, the Trib2 and GAPDH mRNA levels were determined by RT-PCR analysis.
Reverse transcription-PCR (RT-PCR) and quantitative RT-PCR
Total cellular RNA was extracted using the TRIzol (T9424, Sigma-Aldrich) method. For RT-PCR analysis, 2 μg of each RNA aliquot were subjected to cDNA synthesis using 200 U of M-MLV reverse transcriptase and 0.5 μg of oligo (dT) 15 primer (C-1101, Promega). cDNA in 2 μl of the reaction mixture was amplified using 0.5 U of GoTaq DNA polymerase (M8298, Promega). The thermal cycle was as follows: denaturation at 95 °C for 30 s, annealing at 52–58 °C for 30 s depending on the primers used, and extension at 72 °C for 30 s. Each PCR reaction was carried out for 30 cycles, and the PCR products were size-fractionated on a 1.2% agarose gel with ethidium bromide and photographed under UV trans-illumination. Quantitative RT-PCR was performed on an ABI 7500 (Applied Biosystems) sequence detection system with SYBR Green PCR Master Mix (ABS-4309155, Applied Biosystems) according to the manufacturer’s instructions. Experiments were performed in duplicate, and the data were normalized to β-actin mRNA expression. Data were analyzed using the Δ (Δ CT) method and normalized to β-actin. The RT-PCR primer sequences are listed in Supplementary Table S1.
Western blotting
Cell extracts were treated with the appropriate conditions, washed with ice-cold phosphate-buffered saline (PBS), and then lysed in lysis buffer B (20 mM Tris-HCl, 1 mM EGTA, 1 mM EDTA, 10 mM NaCl, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, 30 mM sodium pyrophosphate, 25 mM β-glycerol phosphate, 1% Triton X-100, pH 7.4). Lysates were resolved by SDS-PAGE, transferred onto nitrocellulose membranes, and stained with 0.1% Ponceau S solution (P3504, Sigma-Aldrich). After blocking with 5% nonfat milk, the membranes were immunoblotted with various antibodies, and bound antibodies were visualized with horseradish peroxidase-conjugated secondary antibodies using an enhanced chemiluminescence Western blotting kit (ECL, Amersham Biosciences).
Immunoprecipitation
To immunoprecipitate FLAG-tagged Trib2, TAP-tagged Oct4 and TAP-tagged Sox2 in 293FT cells, the FLAG-tagged Trib2 and TAP-tagged Oct4 constructs were transfected into 293FT cells with the Lipofectamine-plus reagent, and cell lysates were lysed in lysis buffer A. The cell lysates (1 mg) were then incubated with protein G-sepharose (P3296) conjugated with FLAG-agarose beads (F3165) or streptavidin resin for 2 h at 4 °C on a rotating wheel (all from Sigma-Aldrich). The beads were washed extensively with lysis buffer A. Bound proteins were eluted by boiling in 2 × SDS sample buffer and resolved on 10% SDS-PAGE gels for Western blot analysis. Oct4 deletion constructs containing the residues ΔPOU, ΔTAD2, TAD1, N-half, ΔTAD1, POU, ΔPOUH were generated from p3xFLAG-CMV-10-Trib2 using the QuickChange-XL site-directed mutagenesis kit (Stratagene). To immunoprecipitate the FLAG-Oct4 deletion constructs and TAP-Trib2, they were transfected into 293FT cells and then subjected to the same immunoprecipitation protocol as the other constructs.
Purification of recombinant Oct4 and Trib2 proteins and in vitro protein-protein interaction
His-tagged Oct4 and Trib2 maltose-binding protein (MBP) fusion proteins were expressed in Escherichia coli and lysed in lysis buffer B (50 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 0.2% Triton X-100). Recombinant His-Oct4 and MBP-fused Trib2 proteins were purified by affinity chromatography on Ni-NTA agarose (Qiagen Inc., Valencia, CA, USA) or amylose agarose beads (New England Biolabs, Ipswich, MA, USA) according to the manufacturer’s instructions. Purified His-tagged Oct4 and MBP-fused Trib2 proteins were incubated with pull-down buffer. After an additional 1 h of incubation, bound protein complexes were washed four times with binding buffer. The resulting protein complexes were eluted from the beads by boiling in 2 × SDS sample buffer, resolved on SDS-PAGE, and subjected to western blotting with anti-Trib2 antibodies.
Flow cytometry analysis
Oct4-GFP ES and iPS cells were analyzed on a FACS CantoII flow cytometer (BD Biosciences, San Jose, CA, USA). Ten thousand events were acquired on a FACScan flow cytometer (Becton Dickinson, San Jose, CA, USA), and data analysis was performed using CellQuest software (Becton Dickinson, San Jose, CA, USA).
Promoter luciferase assay
293FT cells were seeded on 12-well plates at a density of 1 × 105 cells per well, followed by transfection of the Oct4 promoter in combination with pcDNA3.0-FLAG-Oct4 and p3 × FLAG-CMV-Trib2 using Lipofectamine-plus according to the manufacturer’s instructions. To measure promoter activity in the self-regulatory loop, the CR4 promoter region of the mouse Oct4 promoter, which contains Oct4-binding sites, was cloned by PCR amplification of genomic DNA (forward: 5′-ATGTCTCTTGTCCTGGCCAGTGAGTCACC-3′; reverse: 5′-GCCTCAGCTTCATCGACTTCACCCG-3′). The resulting fragment was subcloned into the KpnI and XhoI sites of the pGL3-basic vector, followed by transfecting 293FT cells with pGL3-basic-CR4 in combination with pcDNA3.0-FLAG-Oc4 and p3xFLAG-CMV-Trib2. Luciferase activity was measured 2–4 days after infection/transfection using the Dual-Luciferase Reporter Assay System and a VICTOR3 Multilabel plate reader (Perkin Elmer, Santa Clara, CA, USA). The pRL-SV40 plasmid served as an internal control for normalizing transfection efficiency.
Chromatin immunoprecipitation
The chromatin immunoprecipitation (ChIP) assay for detecting Oct4 and FLAG-Trib2 in mES cells was performed as described previously. In brief, ESCs were cross-linked with 1% formaldehyde for 10 min, followed by formaldehyde inactivation by incubation with 125 mM glycine and sonication in nuclear lysis buffer (50 mM HEPES, 1 mM EDTA, 140 mM NaCl, 1% Triton X-100, 0.1% SDS). Chromatin extracts containing DNA fragments with an average size of 300 bp were immunoprecipitated using anti-Oct4, anti-FLAG or control antibodies. ChIP DNA fragments were analyzed by qPCR (CR4 region primer sequences: forward 5′-GGAACTGGGTGTGGGGAGGTTGTA-3′; reverse: 5′-AGCAGATTAAGGAAGGGCTAGGACGAGAG-3′).
Production of retroviruses and lentiviruses
Retroviral plasmids carrying reprogramming factors (pMX-OCT4, pMX-SOX2, pMX-KLF4 and pMX-c-Myc) were individually co-transfected with packaging plasmids (gag-pol and VSV-G) into Plat-GP cells using the Lipofectamine-plus reagent. Plat-GP cells (RV-103, Cell Biolabs, Inc., San Diego, CA, USA) were maintained in DMEM with high glucose, 10% FBS, 10 μgml−1 Blasticidin, and 1 × penicillin-streptomycin. To produce lentivirus expressing Trib2, the pLOVE-Trib2 lentiviral expression vector was co-transfected into 293FT cells with packaging plasmids (Δ 8.9 and VSV-G). Virus-containing supernatants were collected 2 days after transfection, passed through a 0.45-μm filter, and stored at −80 °C.
iPS cell generation
1 × 105 MEFs were seeded in individual wells of six-well plates one day prior to infection with retroviruses harboring Oct4, Sox2, Klf4, or c-Myc (1:1:1:1 ratio, Polybrene (10 μgml−1). MEF media was renewed the day after viral infection. On day 2 after infection, MEFs were collected and re-seeded onto Mitomycin C-treated MEFs. The cell media was switched to ES medium 5 days after infection, which was then changed every other day. Cell morphology was monitored under the microscope every day. After counting colonies on the indicated days, the cells were subjected to further analysis.
Alkaline phosphatase (AP) staining and immunofluorescence staining
AP staining was performed for 30 min at room temperature in the dark using the Vector Red AP Substrate Kit I according to the manufacturer's protocol. The cells were incubated with substrate solution at room temperature until suitable staining developed and then photographed with a DFC300FX mounted digital camera (Leica, Solms, Germany). For immunofluorescence staining, mES or iPS cells were fixed in 4% paraformaldehyde in PBS for 10 min, washed twice with PBS, and blocked with 1% FBS in PBS for 30 min; all procedures were performed at room temperature. The fixed specimens were incubated with primary antibodies for 1 h, followed by incubation with secondary antibodies for 1 h. The Oct4 (1:100; Santa Cruz Biotechnology), Sox2 (1:100; Santa Cruz Biotechnology), Nanog (1:100; cell signaling), SSEA-1 (1:100; Santa Cruz Biotechnology), and Trib2 (1:100; Proteintech Group) primary antibodies were detected by Alexa Fluor 488 and Alexa Fluor 568 conjugated secondary antibodies (Thermo Fisher Scientific). The specimens were finally washed and mounted in Vectashield medium with 4′,6-diamidino-2-phenylindole for nuclei visualization. The stained sections were visualized using laser scanning confocal microscopy (FluoView FV1000, Olympus Corporation, Tokyo, Japan).
Teratoma formation and histological analysis
Animal experiments were performed using protocols approved by the Pusan National University Institutional Animal Use and Care Committee. iPS cells were suspended at a density of 1 × 107 cellsml−1 in DMEM and mixed with Matrigel (BD Biosciences). BALB/CA-nu/nu mice were anesthetized with an intraperitoneal injection of 400 mg kg−1 2,2,2-tribromoethanol (Avertin; Sigma-Aldrich). In total, 100 μl of the cell suspension (1 × 106 cells) was subcutaneously injected into the dorsal flanks of the mice. Four weeks after transplantation, teratomas were surgically dissected from the mice. Samples were rinsed once with PBS, fixed in PBS containing 4% formaldehyde, and embedded in paraffin. Sections were stained with hematoxylin and eosin according to standard protocol.
Statistical analysis
Quantitative data are presented as the mean±s.d. from representative experiments (n≥3). The role of chance in the observed differences was tested using unpaired Student’s t-test. Null hypotheses of no difference were rejected if P-values were <0.05.
Results
Trib2 expression is associated with ES cell pluripotency
Tribbles family members share strong sequence similarity in their conserved pseudokinase domain.9 To evaluate the specificity of Trib2, a Tribbles family member, in regulating pluripotency, we examined Trib1, Trib2, Trib3 and Oct4 expression in mES cells, mES cell-derived mEBs, and further-differentiated EBs on day 10 (Diff-d10) using RT-PCR analysis. As shown in Figure 1a, Oct4 and Trib2 expression was similarly decreased during spontaneous mES cell differentiation. However, Trib1 and Trib3 expression was not significantly affected during the differentiation of mESs. Quantitative analysis of the expression levels using real-time PCR showed decreased Trib2 and Oct4 expression during the spontaneous differentiation of mES cells into day 10 differentiated mEBs, whereas the mRNA levels of Trib1 and Trib3 were not changed (Figure 1b). Expression of the differentiation marker Nestin was significantly increased when mES cells differentiated into day 10 mEBs. These results suggest that Trib2 may be implicated in regulating the pluripotent state of mES cells.
Downregulation of Trib2 expression during embryonic stem (ES) cell differentiation. (a) Results of the reverse transcription PCR (RT-PCR) analysis of mouse ES cells (mES) cells, embryonic bodies (EB) and day 10 embryonic bodies (Diff-d10) with probes for Oct4 and the Trib family are shown. The mRNA levels of the Trib family were visualized after 22 cycles of amplification using primers specific for each Trib isoform. (b) The real-time PCR results of mES cells and day 10 embryonic bodies (Diff-d10) with probes for Trib family members, a stem cell marker (Oct4) and a differentiation marker (Nestin) are shown. The experiments were conducted in triplicate and normalized to β-actin. *P<0.05 by Student’s t-test.
Trib2 is required for ES cell pluripotency maintenance
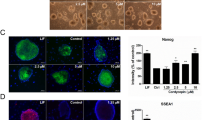
To investigate whether Trib2 is necessary for maintaining the pluripotent phenotype of mES cells, Trib2 expression was knocked down by shRNAs. As shown in Figure 2a, Trib2 shRNA knockdown resulted in morphologically changed mES cells, including reduced colony size and loss of AP activity, which is indicative of differentiation. In addition, when Trib2 expression was knocked down in OG2/ROSA26 mES cells, which express GFP under control of the Oct4 promoter, the GFP-positive cell population was significantly decreased (Figure 2b). Quantitative analysis by flow cytometry showed that the GFP-positive cell population decreased from 50% to 16.9% after Trib2 knockdown. RT-PCR analysis showed that Trib2 knockdown significantly decreased the expression of Oct4, Sox2 and Nanog, whereas the expression of Trib1 and Trib3 did not decrease to the same extent as Oct4 (Figure 2c). Immunocytochemistry analysis showed that Trib2 knockdown substantially decreased the expression of Oct4, Sox2, and SSEA-1 (Figure 2d). To exclude shRNA off-target effects, we performed a rescue experiment. The shRNA used to knock down Trib2 was specific to mouse endogenous Trib2. To perform the rescue experiment, we generated a mES cell line expressing FLAG-tagged human TRIB2. As shown in Figure 2e, knocking down endogenous Trib2 in mES cells decreased the AP-positive colony sizes and numbers. However, ES cells expressing human FLAG-TRIB2 showed little change in their AP-positive colony sizes and numbers after endogenous Trib2 knockdown. The RT-PCR results confirmed successful mouse Trib2 knockdown and human TRIB2 expression (Figure 2e, lower panel). These results strongly suggest that Trib2 expression is required for mES cells to maintain the pluripotent phenotype.
Effect of Trib2 knockdown on the maintenance of mouse embryonic stem (mES) cells. (a) Trib2 expression was knocked down in D3 mES cells. Bright field and alkaline phosphatase (AP) staining images of ES cell colonies from control and Trib2 knockdown cells are shown. Scale bar=50 μm. (b) Trib2 expression was knocked down in Oct4-GFP mES cells. Bright field and fluorescence images of representative colonies from control and Trib2 knockdown cells are shown (left panel). Scale bar=50 μm. Results of the flow cytometry analysis of Oct4-GFP mES cells with or without Trib2 knockdown along with mouse embryonic fibroblasts (MEFs) are shown (right panel). (c) Results of the reverse transcription PCR (RT-PCR)analysis of mES cells with or without Trib2 knockdown with the indicated probes are shown. (d) Fluorescence microscopy images of mES cell colonies with or without Trib2 knockdown are shown after labeling the cells with antibodies against Trib2 (red color) or Oct4 (green color) and 4′-6-Diamidino-2-phenylindole (DAPI, blue color) and overlaid images are shown. Scale bar=20 μm. (e) The rescue of Trib2 knockdown with human TRIB2 (hTRIB2) in mES cells. Mouse Trib2 (mTrib2) was knocked down by mTrib2-specific shRNA in mES cells or mES cells overexpressing hTRIB2, followed by AP staining (upper panel). Scale bar=50 μm. The expression levels of mTrib2, hTRIB2, and GAPDH were detected by RT-PCR analysis on day 4 after infection (lower panel).
Trib2 specifically interacts with Oct4
To examine whether a direct interaction exists between Trib2 and core pluripotency factors, such as Oct4 and Sox2,13, 14 we performed pull-down assays in 293FT cells. After co-transfecting 293FT cells with FLAG-tagged Trib2 and TAP-tagged Oct4 or TAP-tagged Sox2, Oct4 and Sox2 were pulled down with streptavidin beads targeting the TAP tag. As shown in Figure 3a, the Oct4 pull-down co-precipitated Trib2 whereas the Sox2 pull-down did not. Using the opposite approach in which cell lysates were pulled down with anti-FLAG antibodies, Oct4 co-precipitated with Trib2 whereas Sox2 did not (Figure 3b). Immunocytochemistry of mES colonies showed Oct4 and Trib2 co-localization in the nucleus (Supplementary Figure S1). Furthermore, immunoprecipitation with an anti-FLAG antibody in ES cells after FLAG-Trib2 transfection showed that Oct4 co-immunoprecipitated with Trib2 (Figure 3c). These results strongly suggest that endogenous Oct4 and Trib2 interact in ES cells.
Identification of the interaction between Trib2 and Oct4. (a) 293FT cells were transiently transfected with N-terminal streptavidin binding peptide-tagged Oct4 (TAP-Oct4) or N-terminal streptavidin binding peptide-tagged Sox2 (TAP-Sox2) in combination with FLAG-tagged Trib2 (FLAG-Trib2). Results of the western blot analysis of total cell lysates and the precipitates pulled down with the TAP tag after probing with anti-Trib2, anti-Oct4 or anti-Sox2 antibodies are shown. (b) The same cell lysates were pulled down with a FLAG tag, and the western blot analysis results with the anti-Oct4, anti-Sox2 and anti-FLAG antibodies are shown. (c) Interaction of endogenous Trib2 and Oct4 in mES cells. ES cell lysates were subjected to western blot analysis after transfection with FLAG-Trib2 (left panel). FLAG-Trib2 was immunoprecipitated from ES cell lysates, followed by Western blot analysis with the indicated antibodies (right panel). (d) A schematic representation of full-length FLAG-Oct4 and the FLAG-Oct4 deletion constructs along with the Trib2 binding results are shown (upper panel). 293FT cells were transiently transfected with full-length FLAG-Oct4 or each FLAG-Oct4-deletion construct plus Trib2. Results of the western blot analysis of 293FT cell lysates and the precipitates pulled down with an anti-FLAG antibody after probing with anti-FLAG or anti-Trib2 antibodies are shown (lower panel). (e) MBP-Trib2 and MBP-control proteins were expressed in E. coli and immobilized to amylose resin. Purified His-Oct4 was incubated with purified MBP-Trib2 and MBP-control proteins, and the MBP-tagged proteins were precipitated. The level of Oct4 bound to MBP-Trib2 was determined by Western blotting (upper panel). The MBP-Trib2 inputs and the MBP control protein are shown in the lower panel.
Oct4 has POU DNA-binding domains (POUS and POUH) and N-terminal and C-terminal transactivation domains (TAD1 and TAD2).15 To identify the Trib2-interacting regions of Oct4, we utilized a series of Oct4 truncation mutants, as shown in Figure 3d (upper panel).16 After co-transfecting 293FT cells with FLAG-Oct4 or each of the FLAG-Oct4 truncation mutants with TAP-Trib2, they were pulled down with anti-FLAG antibodies. As shown in Figure 3d (lower panel), full-length Oct4 and the ΔTAD2, N-half, ΔTAD1, POU and ΔPOUH truncation mutants, which all contain the POUS domain, interacted with Trib2. The ΔPOU and TAD1 truncation mutants, which contain TAD1 or TAD2 but not the POU domain, did not interact with Trib2. These results suggest that the POUS domain in Oct4 is important for its interaction with Trib2. In addition, when purified His-tagged Oct4 and MBP-fusion Trib2 were incubated in vitro, a MBP-fused Trib2 pull-down co-precipitated His-tagged Oct4, which suggests a direct interaction between Oct4 and Trib2 (Figure 3e). Oct4 is well known to bind Sox2. To examine whether the Trib2 interaction with Oct4 influences the Oct4-Sox2 interaction, we performed an experiment to evaluate the competitive binding of Trib2 and Sox2 to Oct4. 293FT cells were co-transfected with Oct4 in combination with Trib2 and/or Sox2. Individual transfection of Trib2 or Sox2 with Oct4 showed co-precipitation of Trib2 or Sox2 when Oct4 was pulled down (Supplementary Figure S2). However, when Trib2, Sox2, and Oct4 were simultaneously co-transfected, a decrease in Trib2 and Sox2 co-precipitation was observed when Oct4 was pulled down. These results suggest that Trib2 may compete with Sox2 for Oct4 binding. Taken together, these results indicate that the specific and direct interaction between Oct4 and Trib2 could be mediated through the POUS domain in Oct4.
Trib2 positively regulates Oct4 transcriptional activity
Oct4 activates its own promoter.17, 18 To examine whether Trib2 is involved in activating the Oct4 promoter, we performed a transcription reporter assay by co-transfecting NIH3T3 cells with a luciferase reporter driven by a 3 kb upstream region of the Oct4 promoter in combination with Trib2 and Oct4. As shown in Figure 4a, single Trib2 or Oct4 transfection with the reporter increased the Oct4 transcriptional activity significantly. Trib2 and Oct4 co-transfection, however, was synergistic, and dramatically increased the Oct4 transcriptional activity. To further characterize the synergism between Trib2 and Oct4 in regulating Oct4 transcription, we utilized an Oct4 CR4 (conserved region 4)-driven luciferase reporter.16 NIH3T3 cells were co-transfected with the Oct4 CR4-driven luciferase reporter in combination with Trib2 and Oct4. As shown in Figure 4b, Trib2 or Oct4 alone increased the CR4 enhancer activity, and co-transfection of Trib2 and Oct4 increased the CR4 enhancer activity even more. To investigate whether Trib2 occupies the endogenous Oct4 promoter, we examined the binding of Trib2 to the endogenous Oct4 promoter in mES cells. As shown in Figure 4c, a comparison of chromatin immunoprecipitation results using an Oct4 antibody or a FLAG antibody after FLAG-Trib2 transfection to those using control IgGs showed specific binding of Oct4 and Trib2 to the CR4 region of the Oct4 promoter. These results suggest that Oct4 and Trib2 occupy the endogenous Oct4 promoter. Taken together, these results show that Trib2 plays a critical role in regulating Oct4-driven Oct4 transcriptional activity.
The role of Trib2 in regulating the Oct4 promoter activity. (a) NIH3T3 cells were transfected with a 3- kb upstream sequence of the Oct4 promoter-driven luciferase construct in combination with single or double Oct4 and/or Trib2 transfection. The luciferase activities were determined on day 2 after transfection. (b) NIH3T3 cells were transfected with the CR4 region of the Oct4 promoter-driven luciferase construct in combination with single or double Oct4 and/or Trib2 transfection. The luciferase activities were determined on day 2 after transfection. *P<0.05 by Student’s t-test. (c) Chromatin immunoprecipitation (ChIP) analysis of the CR4 region of the endogenous Oct4 promoter. Mouse ES cells expressing FLAG-Trib2 were subjected to ChIP with the indicated antibodies, and qPCR results after normalization to the IgG control are shown. ‘*’ indicates p<0.05 versus the control antibody.
Trib2 regulates the efficiency of somatic cell reprogramming into iPS cells
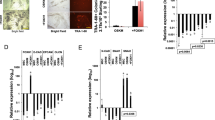
To investigate whether Trib2 contributes to the reprogramming process in the generation of iPS cells from somatic cells, we examined the effect of Trib2 knockdown during the generation of iPS cells from mouse MEFs. MEFs were infected with retroviruses encoding four core transcription factors (Oct4, Sox2, Klf4 and c-Myc) together with lentiviral shRNA against Trib2 (sh-Trib2). As shown in Figure 5a, sh-Trib2 successfully decreased Trib2 expression in MEFs. Introducing Oct4, Sox2, Klf4 and c-Myc (OSKM) into MEFs successfully generated iPS cell colonies, and AP staining on day 28 after infection showed AP-positive colonies. However, Trib2 knockdown along with OSKM significantly decreased the generation of iPS cell colonies and AP-positive colonies (Figure 5b and c). RT-PCR and immunochemical analysis showed that iPS cells generated with OSKM expressed the pluripotency marker genes Oct4, Sox2, Nanog and Klf4 (Figure 5d and e). However, Trib2 knockdown and OSKM significantly decreased the expression of Oct4, Sox2, Nanog and Klf4 in the generated cells (Figure 5d). These results suggest that Trib2 expression is required to reprogram somatic cells into iPS cells. We next evaluated whether the overexpression of Trib2 could enhance reprogramming efficiency. We introduced retroviruses encoding OSKM into Oct4-GFP MEFs with or without lentiviral Trib2 overexpression. As shown in Figure 6a, the lentiviral introduction of Trib2 increased the expression of Trib2 in Oct4-GFP MEFs. When OSKM retroviruses were introduced along with Trib2 overexpression, the AP-positive and GFP-positive iPS cell colony numbers were significantly increased compared to cells transduced with only OSKM (Figure 6b and c). Flow cytometry analysis of reprogramming cells showed that Trib2 overexpression significantly increased the OSKM-induced GFP-positive cell population (Figure 6d). In addition, iPS cells generated from OSKM introduction along with Trib2 overexpression successfully generated teratomas in nude mice (Figure 6e). These results suggest that additional Trib2 expression during the reprogramming process enhances the efficiency of iPS cell generation.
The effect of Trib2 knockdown during the reprogramming of mouse embryonic fibroblasts (MEFs) into iPS cells. (a) The RT-PCR results of MEFs with or without Trib2 knockdown with the indicated probes are shown. Trib2 was visualized after 30 cycles of amplification using Trib2-specific primers. (b) MEFs were reprogrammed into iPS cells by the retroviral transduction of Oct4, Sox2, Klf4 and c-Myc with or without Trib2 knockdown. The alkaline phosphatase (AP) staining of colonies (upper and middle panels) and bright field images of colonies on day 28 after transduction are shown. Scale bar=50 μm. (c) The quantitative analysis of AP-positive colonies is shown. *P<0.05 by Student’s t-test. (d) The RT-PCR results of iPS colonies on day 28 with or without Trib2 knockdown, mES cells and MEFs with the indicated probes are shown. (e) Immunocytochemical images of iPS colonies on day 28 after the retroviral transduction of MEFs with Oct4, Sox2, Klf4 and c-Myc with the indicated antibodies are shown. Scale bar=30 μm.
Effect of Trib2 overexpression during the reprogramming of mouse embryonic fibroblasts (MEFs) into iPS cells. (a) Western blot results of MEFs with or without the lentiviral overexpression of Trib2 with the indicated antibodies. (b) Oct4 promoter-driven GFP MEFs were infected with Oct4, Sox2, Klf4 and c-Myc retroviruses in combination with Trib2-overexpressing lentivirus. The alkaline phosphatase (AP) staining of colonies (upper and middle panels) and fluorescence images of GFP-positive colonies on day 16 after transduction are shown. Scale bar=50 μm. (c) The quantitative analysis of GFP-positive colonies is shown. *P<0.05 by Student’s t-test. (d) The flow cytometry results of iPS colonies on day 16 after Oct4-GFP MEFs were infected with Oct4, Sox2, Klf4 and c-Myc retroviruses in combination with Trib2-overexpressing lentivirus are shown. (e) H&E staining images of teratomas generated from iPS cells with the viral transduction of MEFs with Oct4, Sox2, Klf4, c-Myc and Trib2 are shown. Scale bar=100 μm.
In summary, for the first time, we report that Trib2 is a specific and important factor in the maintenance of mES cell pluripotency. Trib2 shows synergism with Oct4 to positively regulate Oct4 promoter activity. Furthermore, Trib2 is a critical factor during the reprograming of somatic cells into iPS cells. These results will contribute to understanding the mechanisms underlying pluripotency regulation, which may bring clinical regenerative medicine a step closer.
Discussion
In the present study, we demonstrated that Trib2 is an important regulator of ES cell self-renewal and pluripotency induction during the reprogramming process. Unique transcription factors in pluripotent cells form a complex network.17 The expression levels of Trib2 in mES cells and differentiated EBs resemble those of Oct4, an important regulator of pluripotency in undifferentiated ES cells. The pluripotent state-specific gene expression patterns of ES cells can indicate influential clues that maintain their characteristics, such as self-renewal and differentiation potential. Numerous genes whose expression decreases during ES cell differentiation have been identified,19 and several are required to maintain ES cell pluripotency.20, 21 These observations led to our hypothesis that Trib2 is important for maintaining the pluripotent state. We observed that upon shRNA-mediated Trib2 knockdown, ES cells failed to maintain pluripotency. We also observed decreased Oct4, Sox2, Nanog and SSEA-1 expression and abnormal ES cell colony formation in mES cells. These results illustrate the novel aspects of Trib2 in pluripotency induction and maintenance.
We demonstrated a protein-protein interaction between Trib2 and Oct4 using affinity purification methods,22 which allow proteins associated with a target protein to be identified. Oct4 is a master regulator in ES cell development.23 Oct4-interacting proteins are usually identified in human cancers and other diseases by affinity purification, and they possess functional significance related to differentiation, signaling and transcriptional activity.24 Oct4 contains a POU-specific DNA-binding domain (POUS) with a POU homeodomain (POUH) as well as transactivation domains located at its N- (TAD1) and C-termini (TAD2).25, 26 Interestingly, the Oct4 POU domain is an essential regulator in maintaining the self-renewal and pluripotent potential of ES cells.27 In addition, POUH is involved in forming the Oct4-Sox2 complex.28 Our data indicate that the Trib2-interacting region of Oct4 is not observed with other transactivation domains but appears to be specific for the POU domain. Therefore, these results suggest that Trib2 directly interacts with Oct4. Moreover, Trib2 showed a similar binding pattern to Oct4 in the CR4 region (Figure 4c). Trib2 does not have a DNA-binding domain. Therefore, Trib2 likely binds to the CR4 region via binding Oct4, although the molecular mechanism underlying the Trib2-dependent activation of the Oct4 promoter needs to be clarified further.
Our findings in the present study have revealed the essential function of Trib2 in maintaining and establishing ES cell pluripotency. iPS cells closely resemble ES cells in their infinite growth capacity and their ability to differentiate into diverse cell lineages. Investigating the functional proteins and interaction networks involved in the reprogramming process is important to understand the molecular mechanisms underlying pluripotency establishment. Our data demonstrated that inducing Trib2 loss of function by shRNA during the reprogramming process interfered with colony formation, ES cell marker expression and AP activity. In addition, GFP-expressing colonies in which OSKM was introduced and Trib2 was overexpressed were found to have a more efficient reprogramming process than the control. These data suggest that Trib2 regulates reprogramming efficiency in iPS cell generation, implying that Trib2 contributes to the establishment of pluripotency.
Trib2 was identified as an oncogene that induces acute myelogenous leukemia (AML) via a mechanism involving the downregulation of C/EBPα.29 Among Tribbles family members, Trib1 and Trib2 induced AML from hematopoietic stem cells whereas Trib3 did not.30 To reprogram B cells into iPS cells with inducible OSKM expression, ectopic C/EBPα expression was necessary.31 While reprogramming mouse primary B cells by OSKM, transient C/EBPα expression with Oct4 expression increased the efficiency of iPS cell generation by 100-fold in 2 days, suggesting that C/EBPα functions as a ‘path-breaker’.32 A recent report showed that in AML, the elevated expression of C/EBPα p42 and left p30, an oncogenic isoform, and the complete absence of C/EBPα expression abrogated the induction of AML by Trib2.33 In melanoma, TRIB2 acts as an oncogene by suppressing FOXO, which regulates the identity of human ES cells.21, 34 Though the detailed molecular mechanism of how Trib2 contributes to the reprogramming process currently remains elusive, our findings suggest that Trib2 is a critical factor in maintaining and inducing pluripotency.
In summary, the novel aspect of our study is that Trib2 maintains pluripotency and progresses cellular reprogramming. Trib2 expression was downregulated in differentiated ES cells, and Trib2 depletion reduced the pluripotency of ES cells. In support of these results, we demonstrated that Oct4, an essential regulator of pluripotency, directly interacts with Trib2, and its interaction increases Oct4 promoter activity. Finally, Trib2 facilitates iPS cell generation through the Trib2/Oct4 complex.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Niwa H . How is pluripotency determined and maintained? Development 2007; 134: 635–646.
Jaenisch R, Young R . Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 2008; 132: 567–582.
Takahashi K, Yamanaka S . Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126: 663–676.
Hanna JH, Saha K, Jaenisch R . Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell 2010; 143: 508–525.
Stadtfeld M, Hochedlinger K . Induced pluripotency: history, mechanisms, and applications. Genes Dev 2010; 24: 2239–2263.
Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R . Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 2003; 17: 126–140.
Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 2003; 113: 631–642.
Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 1998; 95: 379–391.
Hegedus Z, Czibula A, Kiss-Toth E . Tribbles: novel regulators of cell function; evolutionary aspects. Cell Mol Life Sci 2006; 63: 1632–1641.
Wilkin F, Suarez-Huerta N, Robaye B, Peetermans J, Libert F, Dumont JE et al. Characterization of a phosphoprotein whose mRNA is regulated by the mitogenic pathways in dog thyroid cells. Eur J Biochem 1997; 248: 660–668.
Grandinetti KB, Stevens TA, Ha S, Salamone RJ, Walker JR, Zhang J et al. Overexpression of TRIB2 in human lung cancers contributes to tumorigenesis through downregulation of C/EBPalpha. Oncogene 2011; 30: 3328–3335.
Zanella F, Renner O, Garcia B, Callejas S, Dopazo A, Peregrina S et al. Human TRIB2 is a repressor of FOXO that contributes to the malignant phenotype of melanoma cells. Oncogene 2010; 29: 2973–2982.
Kim J, Chu J, Shen X, Wang J, Orkin SH . An extended transcriptional network for pluripotency of embryonic stem cells. Cell 2008; 132: 1049–1061.
van den Berg DL, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J et al. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 2010; 6: 369–381.
Pesce M, Scholer HR . Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells 2001; 19: 271–278.
Do EK, Cheon HC, Jang IH, Choi EJ, Heo SC, Kang KT et al. Reptin regulates pluripotency of embryonic stem cells and somatic cell reprogramming through Oct4-dependent mechanism. Stem Cells 2014; 32: 3126–3136.
Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008; 133: 1106–1117.
Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 2006; 38: 431–440.
Brandenberger R, Wei H, Zhang S, Lei S, Murage J, Fisk GJ et al. Transcriptome characterization elucidates signaling networks that control human ES cell growth and differentiation. Nat Biotechnol 2004; 22: 707–716.
Lim LS, Loh YH, Zhang W, Li Y, Chen X, Wang Y et al. Zic3 is required for maintenance of pluripotency in embryonic stem cells. Mol Biol Cell 2007; 18: 1348–1358.
Zhang X, Yalcin S, Lee DF, Yeh TY, Lee SM, Su J et al. FOXO1 is an essential regulator of pluripotency in human embryonic stem cells. Nat Cell Biol 2011; 13: 1092–1099.
Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B . A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 1999; 17: 1030–1032.
Palmieri SL, Peter W, Hess H, Scholer HR . Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev Biol 1994; 166: 259–267.
Pardo M, Lang B, Yu L, Prosser H, Bradley A, Babu MM et al. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell 2010; 6: 382–395.
Brehm A, Ohbo K, Scholer H . The carboxy-terminal transactivation domain of Oct-4 acquires cell specificity through the POU domain. Mol Cell Biol 1997; 17: 154–162.
Scholer HR, Ruppert S, Suzuki N, Chowdhury K, Gruss P . New type of POU domain in germ line-specific protein Oct-4. Nature 1990; 344: 435–439.
Niwa H, Masui S, Chambers I, Smith AG, Miyazaki J . Phenotypic complementation establishes requirements for specific POU domain and generic transactivation function of Oct-3/4 in embryonic stem cells. Mol Cell Biol 2002; 22: 1526–1536.
Ambrosetti DC, Scholer HR, Dailey L, Basilico C . Modulation of the activity of multiple transcriptional activation domains by the DNA binding domains mediates the synergistic action of Sox2 and Oct-3 on the fibroblast growth factor-4 enhancer. J Biol Chem 2000; 275: 23387–23397.
Keeshan K, He Y, Wouters BJ, Shestova O, Xu L, Sai H et al. Tribbles homolog 2 inactivates C/EBPalpha and causes acute myelogenous leukemia. Cancer Cell 2006; 10: 401–411.
Dedhia PH, Keeshan K, Uljon S, Xu L, Vega ME, Shestova O et al. Differential ability of Tribbles family members to promote degradation of C/EBPalpha and induce acute myelogenous leukemia. Blood 2010; 116: 1321–1328.
Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell 2008; 133: 250–264.
Di Stefano B, Sardina JL, van Oevelen C, Collombet S, Kallin EM, Vicent GP et al. C/EBPalpha poises B cells for rapid reprogramming into induced pluripotent stem cells. Nature 2014; 506: 235–239.
O'Connor C, Lohan F, Campos J, Ohlsson E, Salome M, Forde C et al. The presence of C/EBPalpha and its degradation are both required for TRIB2-mediated leukaemia. Oncogene 2016; 35: 5272–5281.
Gonzales KA, Ng HH . FoxO: a new addition to the ESC cartel. Cell Stem Cell 2011; 9: 181–183.
Acknowledgements
This study was financially supported by the 2015 Post-Doc Development program of Pusan National University and Basic Science Research Programs through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (2015R1D1A4A01019579), the MRC program (NRF-2015R1A5A2009656), and the Ministry of Science, ICT & Future Planning (2015R1C1A2A01053377).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflicts of interest.
Additional information
Supplementary Figures and Table accompanies the paper on Experimental & Molecular Medicine website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Do, E., Park, J., Cheon, H. et al. Trib2 regulates the pluripotency of embryonic stem cells and enhances reprogramming efficiency. Exp Mol Med 49, e401 (2017). https://doi.org/10.1038/emm.2017.191
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/emm.2017.191
This article is cited by
-
Tribbles homolog 2 (Trib2), a pseudo serine/threonine kinase in tumorigenesis and stem cell fate decisions
Cell Communication and Signaling (2021)
-
TRIB2 functions as novel oncogene in colorectal cancer by blocking cellular senescence through AP4/p21 signaling
Molecular Cancer (2018)