Abstract
A small set of gastric adenocarcinomas (9%) harbor Epstein–Barr virus (EBV) DNA within malignant cells, and the virus is not an innocent bystander but rather is intimately linked to pathogenesis and tumor maintenance. Evidence comes from unique genomic features of host DNA, mRNA, microRNA and CpG methylation profiles as revealed by recent comprehensive genomic analysis by The Cancer Genome Atlas Network. Their data show that gastric cancer is not one disease but rather comprises four major classes: EBV-positive, microsatellite instability (MSI), genomically stable and chromosome instability. The EBV-positive class has even more marked CpG methylation than does the MSI class, and viral cancers have a unique pattern of methylation linked to the downregulation of CDKN2A (p16) but not MLH1. EBV-positive cancers often have mutated PIK3CA and ARID1A and an amplified 9p24.1 locus linked to overexpression of JAK2, CD274 (PD-L1) and PDCD1LG2 (PD-L2). Multiple noncoding viral RNAs are highly expressed. Patients who fail standard therapy may qualify for enrollment in clinical trials targeting cancer-related human gene pathways or promoting destruction of infected cells through lytic induction of EBV genes. Genomic tests such as the GastroGenus Gastric Cancer Classifier are available to identify actionable variants in formalin-fixed cancer tissue of affected patients.
Similar content being viewed by others
Introduction
Epstein–Barr virus (EBV) is present within the malignant cells in ~9% of all gastric adenocarcinomas worldwide.1 Another pathogen, Helicobacter pylori (Hp), commonly present in the stomach, confers a fivefold increased cancer risk.2 Gastric cancer is the leading cause of infection-related cancer mortality and is projected to soon rise to rank eighth in all-cause mortality.3, 4 By the time gastric cancer is diagnosed, a billion or more malignant cells are typically present harboring dozens to thousands of somatic gene variants. Genomic tests show promise to identify driver mutations such as ERBB2 amplification for which targeted therapy may be effective.5 Understanding the functional impact of genomic variation promotes selection of drugs that target relevant pathways and thus could overcome unwanted effects that promote growth, thwart apoptosis, elude the immune system or impair DNA repair mechanisms that foster more mutagenesis.6, 7, 8, 9
Enrollment in clinical trials of targeted therapy increasingly relies on results of tests for pertinent genes and gene products. This article describes the current state of genomic assay development for gastric cancer and the opportunities to capitalize on EBV and its effectors as targets for therapy.
Four major molecular classes of gastric adenocarcinoma
Data from The Cancer Genome Atlas (TCGA) Network suggest that gastric adenocarcinoma is not one disease but rather comprises four molecular classes, as discovered by comprehensive tests of 295 frozen gastric cancer tissues from untreated patients using whole exome sequencing, RNAseq, microRNA sequencing, SNP array, methylation array, reverse-phase protein array, microsatellite instability (MSI) testing and (in 107 tumors) whole genome sequencing.10 Matched germline DNA (generally from blood) and non-malignant gastric mucosa were also examined. The summary of key findings is provided in Table 1.
MicroRNA and messenger RNA profiles in EBV-positive gastric adenocarcinoma
EBV was the first virus recognized to encode its own microRNAs. MicroRNA and mRNA profiles are achievable in formalin-fixed, paraffin-embedded tissue using massively parallel sequencing or array technology. The summary of RNAs distinguishing each of the four cancer classes from non-malignant gastric mucosa is provided in Table 2.10, 11, 12, 13, 14, 15, 16, 17, 18 Several downregulated mRNAs are shared among the four gastric cancer classes, implying these RNAs are pancancer markers in the stomach. Conversely, CST1 is upregulated in all four cancer classes.
EBV-associated DNA hypermethylation
A striking feature of EBV-positive gastric cancer is extreme CpG hypermethylation, including both promoter and non-promoter CpG islands of the human genome.10, 19, 20 Notably, the pattern of methylation is even more extensive than the classic CpG island methylator phenotype observed in the MSI class of gastric cancers, and is more extensive than was seen in any tumor type previously studied by the TCGA Network.10, 21, 22 Furthermore, EBV and MSI methylation patterns are distinct, with EBV-positive tumors displaying CDKN2A (p16) promoter hypermethylation but lacking MLH1 hypermethylation.10, 23, 24
The genes silenced in virtually all EBV-positive cancers in concert with promoter hypermethylation are listed in Table 3.10 The RCOR2 gene exhibited methylation-related silencing in 100% of EBV-positive and in 0% of EBV-negative gastric cancers. Work on cell lines suggests that downregulation of the RCOR2 transcription factor promotes hypermethylation, whereas expression of RCOR2 promotes reprogramming to stem cell pluripotency.25, 26
EBV infection leads to extensive methylation of both host and viral genomes, providing a mechanism for viral control of cellular functions promoting viral persistence and propagation.20, 27, 28, 29, 30, 31, 32 EBV BZLF1 was the first protein ever shown to preferentially bind methylated promoters to induce gene expression, thus overcoming transcriptional silencing to switch an infected cell from viral latency to active, lytic viral replication. Interestingly, Hp infection is also associated with hypermethylation.33 In vitro evidence points to demethylating drugs that can reverse the effect, but clinical trials of EBV-positive tumors (lymphoma and nasopharyngeal carcinoma) treated with 5-azacytidine plus phenylbutyrate had disappointing results.34
EBV-directed therapy, and genomic tests to monitor efficacy
As viral DNA and selected viral gene products (see below) are present in every malignant cell of an EBV-positive tumor, a compelling cure for cancer would be to eliminate all infected cells. Strategies for virus-directed therapy are listed in Table 4.
Lytic induction therapy is a rational means to promote destruction of infected cells, and the putative mechanisms of action were recently summarized by Kenney and Mertz.35 Radiation therapy and selected drugs induce lytic viral gene expression, enhancing immune recognition of foreign proteins.35, 36, 37 Radiation is effective in treating some EBV-infected cancers such as nasopharyngeal carcinoma and Hodgkin’s lymphoma. Histone deacetylase inhibitors are among the most potent inducers of active viral replication.38 Short-chain fatty acids such as butyrate are also good inducers having reasonable safety profiles. Butyrate is produced naturally by certain bacteria comprising the oral and gastric flora.39 Nucleoside analog drugs such as gancyclovir may enhance cell death during lytic induction therapy35 (see Figure 1). Two clinical trials reported positive biologic effects and minimal toxicity.40, 41
During latent infection, a very limited repertoire of viral gene is expressed. However, when an infected cell switches into the lytic phase of active viral replication, dozens of viral proteins are expressed that trigger immune recognition and destruction. Cell death may be enhanced by administering the anti-viral drug gancyclovir, a purine analog that is incorporated into DNA strands by DNA polymerase. Viral thymidine kinase (BXLF1) and serine/threonine protein kinase/phosphotransferase (BGLF4) phosphorylate gancyclovir, which then stalls DNA synthesis and triggers apoptosis. Intercellular transfer of phosphorylated gancyclovir can result in death of adjacent cells.
A major aim of the lytic induction therapy is to provoke host cell expression of immunogenic foreign proteins that incite immune responses. Decades of experience treating EBV-driven posttransplant lymphoproliferation shows that cutting back on iatrogenic-immunosuppressive drugs restores the body’s natural ability to control EBV infection, potentially reducing tumor burden as reflected by lower viral load in the plasma.42
In solid tumor patients, EBV-directed T cells infused in concert with immune modulators have some efficacy against infected cancers.43 Drugs that were ineffective as single agents are now being considered in combination, such as a histone deacetylase inhibitor (e.g. vorinostat, suberoylanilide hydroxamic acid or valproate) plus the proteasome inhibitor bortezomib.35, 44, 45, 46, 47, 48, 49 Triple drug therapy with gancyclovir plus gemcitabine and valproate (both of which induce lytic viral replication) showed anecdotal value in stabilizing three nasopharyngeal carcinoma patients.41 Clinical trials should report EBV status as well as pertinent genomic features to characterize exceptional responders and to shed light on mechanisms of action in relevant biochemical pathways.
Laboratory tests can measure the degree of lytic induction using quantitative PCR to quantify viral genomes and transcriptome profiles to measure lytic mRNAs. Parallel tests of human RNA evaluate the impact on pertinent cellular biochemical pathways. These tests may be applied to biopsy material, although periodic rebiopsy is impractical and risky. Plasma is emerging as a more practical specimen type in which to measure tumor markers (EBV viral load, somatic mutations and microRNA or methylation profiles) to assess near-term effects of intervention and long-term tumor burden.50, 51, 52, 53
Host gene mutation
A salient feature of EBV-positive gastric cancer is PIK3CA mutation, which is found in 80% of such cancers compared with only 3–42% for cancers in the other three molecular classes. Interestingly, nearly half of EBV-positive nasopharyngeal carcinomas harbor PIK3CA mutation, implicating a common theme to viral carcinogenesis despite squamous vs glandular histologies. This is a classic example of genomic features linking two cancer types despite different histologies and anatomic sites.54
In EBV-positive gastric cancer, PIK3CA mutations are not restricted to hotspots (helical or kinase domains) but rather are spread across many gene segments.10 Clinical trials of various PIK3CA/AKT/mTOR inhibitors (e.g. everolimus, BEZ235) showed disappointing results as single agents in gastric cancer, and dual pathway inhibition is now being explored.55, 56, 57, 58, 59, 60 Unique cancer prevention strategies are also being explored for PIK3CA-mutated cancers.
Human genes commonly mutated in gastric cancer are listed in Table 5.10 The vast majority of cancers have mutation or gene amplification in a targetable signaling pathway (receptor tyrosine kinases such as MET or EGFR, JAK/STAT, GTPase (RHO/RAS/RAF), PIK3CA/MTOR/PTEN, CTNNB1). RNAseq revealed MET exon skipping (of exons 2, 18/19 or 19) associated with overexpression of the encoded receptor. MET gene amplification was found in about a third of EBV-positive cancers.
EBV-positive cancers typically lack TP53 mutation, although TP53 was nearly always mutated in ‘chromosome instability’ cancers,10 which might be detectable by immunohistochemical evidence of TP53 protein accumulation.61 Indeed, TP53 immunostains already serve as an adjunct to histopathology in predicting progression of Barrett’s lesions to cancer.62 Shimizu et al.63 recently reported TP53 or ARID1A mutation at low allele frequency in non-malignant gastric mucosa of patients with Hp infection, and Hp-related mutagenesis is purportedly related to activation of cytidine deaminase leading to characteristic C>T transversions of GpCpX motifs.
EBV-related immune system dysfunction identified in TCGA studies
Autocrine or paracrine growth factors seem to promote growth of tumor cells. The TCGA Network reported that, compared with EBV-negative cancers, EBV-positive cancers have evidence of hyperactive adaptive and innate immunity, with evidence of T-cell activation via the cytokines interleukin-2 (IL-2), IL-12, IL-23 and IL-27.10 Some T-cell activation evidence undoubtedly emanates from tumor-infiltrating lymphocytes that tend to be abundant in infected cancer tissues, yet are unable to control growth of infected tumor cells.64 Other features of EBV-positive cancers are (1) diminished glucocorticoid signaling, suggesting an opportunity to test dexamethasone and other immune modulators, (2) defective cell adhesion and (3) strong caspase activity that might be exploited to tip the balance towards death of infected cells.
When compared with non-malignant gastric mucosa, EBV-positive cancers exhibit many of the same biochemical features as uninfected cancers, including strong DNA damage response pathways. A major difference is that interferon-γ and IFN-γ-induced interferon regulatory factor-1 in the IL-2-STAT4 pathway are overexpressed in EBV-positive cancers versus uninfected cancers.65
EBV-related receptor kinase signaling
Compared with non-malignant mucosa, TCGA investigators reported that the two most marked features of EBV-positive cancers are diminished hypoxia-inducible factor 1α-related activity and diminished ERBB receptor signaling. These findings raise the question of whether angiogenesis inhibitors or ERBB family inhibitors might have differential efficacy in infected vs uninfected cancers. Pending further studies, it seems reasonable to continue to follow customary clinical recommendations66 regarding use of drugs such as ramucirumab targeting vascular endothelial growth factor receptor-2 involved in angiogenesis67, 68, 69 or trastuzumab in the setting of ERBB2 (HER2) overexpression/amplification.
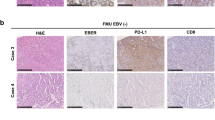
EBV-positive cancers have evidence of activated BMP (bone morphogenetic protein) signaling, implicating that the BMP/SMAD pathway as a potential therapeutic target. In addition, potentially druggable JAK2 or MET gene amplifications are relatively frequent among EBV-positive cancers.7 Intriguing studies show EBV-positive gastric cancers preferentially overexpress CD274 and PDCD1LG2 (PD-L1 and PD-L2) as revealed by RNAseq and by protein localization to malignant cells by immunohistochemistry.10, 70 Prior studies of lymphoma likewise revealed EBV-associated upregulation of CD274 on the cell surface, which is IFN-γ-mediated, and thus inhibits killing of infected cells by cytotoxic T cells expressing PD-1 ligand.71 Gain of four or more copies of the CD274 and PDC1LG2 genes, or mutation in the 3′-untranslated region of CD274, are alternative mechanisms of overexpression.72 PDCD1LG2 thwarts T-helper type 2 T-cell function. Further work is needed to explore virus-related immune evasion mechanisms, particularly now that PD/PD-L inhibitor drugs are available.
Regardless of EBV status, most gastric cancers have at least one aberrancy in a druggable pathway such as receptor tyrosine kinase signaling.10, 73, 74, 75, 76, 77, 78, 79 Newly identified in ‘genomically stable’ gastric cancers is activated RHOA singaling via mutation of RHOA GTPase, or fusion events in RHOA inhibitors (ARHGAPs), broadening the opportunity to test inhibitors of the RHOA effector ROCK that are well studied in vascular biology. ROCK functions as a serine/threonine kinase impacting CDH1-mediated cell adhesion, tumor microenvironment and actin structural biology.10, 80, 81 CDH1, RHOA and ARHGAP defects are common and are mutually exclusive. Hp infection reportedly activates members of the RHO family of GTPases.
The CD44 gene is preferentially amplified in EBV-positive cancers, and other commonly amplified or deleted genes are listed in Table 6. Previously identified were a CD44 protein variant or a CD44-SLC1A2 or SLC34A2-ROS1 fusion in cancer tissues.82, 83 Fusion genes are technically difficult to find by next-generation sequencing, and expensive to identify by fluorescence in situ hybridization. However, once identified as a somatic variant, a fusion gene is considered to be a particularly good tumor marker in that it tends to be quite specific for neoplastic cells and is identifiable in ‘discarded’ next-generation sequencing reads that do not align to the reference sequence using standard bioinformatic tools.84 Low-level translocation detected by quantitative PCR could serve as a marker of minimal residual disease.
EBV gene expression
Among 87 EBV-encoded mRNAs, 15 are highly expressed in the majority of EBV-positive gastric cancers according to the TCGA RNAseq data. Eight of these EBV transcripts (BARF0, BALF3, BALF4, BALF5, A73, RPMS1, LF2 and LF1) are encoded in the BamH1A region of the viral genome where the expressed viral miRs are also encoded.10, 65, 85 Although the medical literature suggests that latent membrane protein 1 is infrequently expressed in infected gastric adenocarcinoma by protein assays, TCGA RNAseq revealed that both EBV latent membrane protein encoding mRNAs (LMP1, LMP2A) were consistently expressed, albeit at low level. Latent membrane protein 2 or other viral gene products may contribute to hypermethylated DNA that is characteristic of infected cancers.86
Another interesting finding is consistent expression of EBV BNLF2a, which acts to inhibit the transporter associated with antigen processing, potentially thwarting antigen presentation.87 Also consistently expressed is the BILF1 G-protein-coupled receptor that downregulates human leukocyte antigen class 1 protein expression.88 Both BNLF2a and BILF1 are thus involved in the cellular evasion of immune destruction, and are associated with the upregulation of the druggable natural killer/T-cell inhibitor IDO1 (indoleamine 2,3-dioxygenase 1) in EBV-positive tumors.65 As the IDO1 enzyme depletes tryptophan, it raises the question whether foods rich in tryptophan could overcome the effect. A viral gene of uncertain function, BNLF2b, is highly expressed, whereas EBNA1 and LF3 are expressed consistently but at low levels. The remaining latent and lytic viral genes (including the other EBNAs) are expressed at very low levels or only in a fraction of tumors.10
Selected viral microRNAs (encoded in the viral BamH1A region of the viral genome) were highly expressed in all infected cancers in a fairly consistent pattern, according to the TCGA microRNA sequencing data.10 The most highly expressed of these EBV microRNAs are BART 10-5p, 7-3p and 7-5p. Emerging data reveals their impact on gastric carcinogenesis.89, 90, 91
EBV genome structure and a rare chromosomal integration event
The EBV genome persists as an episome (double-stranded DNA of almost 172 kb) inside latently infected cells, with variable numbers of terminal repeat sequences within the circularized viral genome serving as a marker of clonality.92 The presence of clonal viral DNA in EBV-positive gastric cancers is evidence that infection precedes malignant transformation, a concept that is reinforced by histochemical evidence of EBER expression in all neoplastic cells of a given patient, and by evidence of EBV in some dysplastic lesions of the stomach mucosa.93 Whether EBV infection of epithelial cells occurs early or late during carcinogenesis is a subject of ongoing investigation.94, 95, 96
Multiple viral polymorphisms have been reported,97, 98, 99, 100 and the extent to which these mutations in the EBV genome impact gastric carcinogenesis is unknown. In the TCGA study, one gastric cancer had evidence of integration of the EBV genome into the human genome as revealed by multiple independent RNAseq reads revealing a fusion transcript predicted to join the first 20 amino acids of the human plasminogen receptor (PLGRKT, alias C9orf46) to almost the entire coding sequence of the early lytic EBV gene BHLF1 (alias EA-D). In this particular tumor, BHLF1 was expressed but the remaining viral gene expression pattern was not markedly different from other infected tumors. It should be noted that PLGRKT is located alongside JAK2, CD274 (PD-L1) and PDCD1LG2 (PD-L2) within the 9p24.1 locus that is commonly amplified in EBV-positive gastric cancers. Overall, the findings imply that viral integration can occur, is rare event or else does not does not frequently result in fusion transcripts, and may contribute to the activation of known oncogenes.
Histopathology of EBV-positive gastric adenocarcinoma tissues
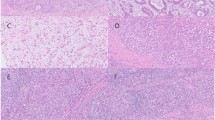
Prior work has shown that carcinoma with lymphoid stroma is characteristic of many EBV-positive cancers, although some infected cancers have more conventional tubular (intestinal) appearance by microscopy. Histologic classification of cancers using Lauren or World Health Organization schemes reveals that ‘genomically stable’ cancers are enriched for diffuse histology10 (see Table 7). Further work should delve into histologic features differentiating the four molecular classes such as the character of lymphoid stroma and epithelial cell cytology/architecture (e.g. signet ring cell type, solid type vs isolated cell diffuse architecture as distinguished by Carneiro et al.101).
In EBV-positive cancers, the EBV genome is localized within the nucleus of malignant cells and is propagated to daughter cells during cell division. EBER in situ hybridization is considered the gold standard assay for assigning a tumor as ‘EBV-positive’ by virtue of localization of abundantly expressed EBER1 or EBER2 long noncoding RNAs to malignant cells.92 High levels of EBV DNA (using quantitative PCR) or encoded viral RNA (using microarray, RNAseq or microRNA sequencing) are surrogate methods to distinguish EBV-positive cancers from EBV-negative cancers.1, 10, 102 High levels of EBV DNA are also seen during active viral infection (e.g. infectious mononucleosis, chronic active EBV infection, oral hairy leukoplakia), whereas low-level infection is commonly present in adult humans who were previously infected and then carry the virus for life in a small proportion of B lymphocytes.92
Clinicopathologic features impacting treatment
From a clinical management perspective, it is clear that factors independent of molecular class impact treatment decisions at this time. Tumor stage is critical for devising plans for surgical resection, radiation and chemotherapy. ERBB2 (HER2) gene amplification qualifies metastatic cancer patients for HER2-directed antibody therapy.103 Yet, tumor stage, ERBB2 status and survival were not strongly linked with molecular class,10 emphasizing that molecular class represents a conceptual biologic framework more than a practical tool at this time. Much remains to be learned about what appears to be four major routes of disease pathogenesis, and to capitalize on that understanding for purposes of diagnosis, management and prevention of cancer. Meanwhile, as most cancers have somatic variants in biochemical pathways that are putative targets for existing drugs, design of clinical trials is warranted to test the efficacy of targeted therapies.
Implications for surgical pathologists and molecular pathologists
The reason why pathologists classify cancers is to promote clinical decision-making that improves patient outcomes. Classification criteria are periodically revised to incorporate new scientific evidence and methods of analysis. At this time, assignment to one of four molecular classes does not appear to add value beyond the features already actionable for clinical decision-making in gastric tumors (e.g. carcinoma vs gastrointestinal stromal tumor, ERBB2 (HER2) gene amplification in metastatic carcinoma, stage). Nevertheless, microscopic and ancillary tools can certainly be validated and applied to distinguish the four molecular subtypes at a reasonable cost, making it feasible to retrospectively and prospectively test clinical trial cohorts for molecular class.
The GastroGenus Gastric Cancer Classifier is a genomic assay that tests for EBV positivity, MLH1 promoter methylation and sequences of selected cancer genes in the context of histopathologic classification. Results of the test in paraffin-embedded tumor tissue are interpreted with respect to molecular class and to find actionable gene variants in patients who fail standard therapy.104 Although therapy options for metastatic cancer patients remain limited and largely palliative,103 clinicians and patients increasingly demand genomic data by which to evaluate options for clinical trial enrollment, the results of which are likely to enhance understanding of pathogenesis and define the role of clinicopathologic tests in predicting drug efficacy.
Localization of EBV to malignant cells by EBER in situ hybridization remains the gold standard for assigning the EBV-positive class of cancers.92 EBV-positive gastric cancer patients qualify for enrollment in clinical trials of EBV-directed therapy (e.g. NCT00982449, NCT02080416). These trials use lytic induction therapy to convert infected cells from latent to replicative phases of viral infection, which is hypothesized to trigger cell death with potential for bystander killing of adjacent cells.35, 48
Also available are multiple trials of targeted therapy for gastric cancer having a particular gene defect or gene expression status. Among these trials are dozens for ERRB2 (HER2) gene defects (e.g. NCT01602406); NCT01613950 for PIK3CA mutation; NCT02016534 or NCT01874938 for MET gene amplification, NCT02052778 for FGF or FGFR gene defects; NCT02187783 for CDK4, CDK6, CCND1, CCND3 or CDKN2A defects; NCT02022982 for KRAS mutation; and NCT01522820 for CTAG1B or CTAG2 expression.
It is likely that the MSI molecular class of gastric cancers can be identified by virtue of MSI-high status (which requires testing of both tumor and germline DNA) or by MLH1 hypermethylation testing that requires only tumor tissue. Heretofore, MSI status has not generally been evaluated in gastric cancer patients except in the rare instance of suspected Lynch syndrome (e.g. personal or family history of pertinent cancer types, young age at diagnosis, blood relative with Lynch syndrome). The MSI test is used primarily to identify evidence of DNA mismatch repair defects in colorectal or endometrial cancer patients as part of Lynch syndrome screening.105 With the advent of massive parallel sequencing panels to identify germline variants in mismatch repair genes, it is likely that the MSI test will eventually be replaced by more cost-effective and direct means of testing for heritable cancer risk.106, 107, 108 For purposes of assigning molecular class of gastric cancer, MLH1 methylation testing is likely to be a suitable surrogate test. A treatment trial of PD-1 antibody therapy (pembrolizumab, NCT01876511) currently enrolls patients with ‘MSI-positive’ gastric cancer. Methylation-related silencing of MLH1 undoubtedly contributes to defective DNA repair and accumulating mutations in cancer tissue. MLH1 promoter methylation also suggests better response to fluorouracil chemotherapy.
Heritable predisposition to gastric cancer
Familial predisposition to gastric cancer is being examined in a clinical trial of subjects from high-risk families (NCT00172861). Pertinent hereditary cancer syndromes include Peutz-Jeghers (STK11), Li-Fraumeni (TP53), Lynch (MLH1, MSH2, MSH6, PMS2, EPCAM, MUTYH) and Cowden (PTEN) syndromes, as well as hereditary diffuse gastric cancer (CDH1).109 In the latter disorder, a hypomethylating drug is being considered to prevent cancer progression from premalignant lesions by thwarting CDH1 promoter methylation. Table 8 lists genes reportedly associated with gastric cancer predisposition, some of which encode immune response factors that defend against EBV, Hp and other pathogens.
Clinical implementation of genomic services
Well-validated genomic tests can provide robust, accurate and reproducible results that are powerful by virtue of the number of analytes that are evaluated, redundancy that boosts confidence in results, cross-species flexibility to detect pathogen-related human disease and a growing evidence base linking genetic findings to disease status in a manner that promotes favorable patient outcomes. Genomic tests typically analyze dozens to billions of targets in a single assay using microarrays or massive parallel sequencing. Performance studies of molecular technology have shown that genomic profiles of DNA, mRNA, methylation status and noncoding RNA can be rendered analytically sound and clinically informative for medical decision-making in clinical trials and ultimately in routine patient care.110, 111, 112, 113, 114, 115, 116
Building on decades of well-honed principles of laboratory medicine, pathologists and other laboratory professionals provide genomic services in clinical settings.111 Implementation is supported by (1) reliable commercial sources of reagents, supplies, instruments and software; (2) advances in biospecimen science promoting integrity of the nucleic acids input to the assays;117 (3) novel quality control reagents and methods to judge assay performance;111, 118 (4) informatics to facilitate interpretation of complex data generated from assays of patient and control specimens; (5) evidence of analytic and clinical performance; and (6) justification that the assay adds value (e.g. is faster, cheaper or more informative) than current means of diagnosis, monitoring, preventing or predicting efficacy of a given intervention in the pertinent population.
Each testing laboratory has a ‘standard operating procedure’ that, along with a ‘validation report’, establishes the evidence base substantiating indications for testing, specimen collection and processing, step-by-step analysis, quality control processes and guidance for interpreting data and for generating a report to the patient’s medical record.119 Analytic interpretation is the process by which raw data is converted into reportable results. Clinical interpretation is the process by which a pathologist or other laboratory professional judges the medical significance of results in light of the clinical indication for which the service was ordered, the findings and the intended use of test results.
Analytic interpretation relies on thorough understanding of the technical strengths and weaknesses of the test system based on prior experience gathered during validation studies, literature review and subsequent clinical practice. Owing to test complexity and the need to interpret results in the context of the many quality assurance measures that are in place (including, in the case of molecular oncology specimens, histopathologic evaluation of the input lesion), it is clear that the resulting genomic sequence is not a stand-alone feature but rather a component of a package of technical and professional work constituting a medical service.
Interpretation is impacted by the level of confidence in results, as revealed by redundant findings (e.g. depth of coverage, mutant allele frequency, multiple tests of the same analyte or pathway, presence of both 5′ and 3′ ends of an mRNA) and the outcome of controls and quality checks (e.g. endogenous controls to evaluate adequacy of patient nucleic acid for the intended use, exogenous and spiked controls to evaluate run- and specimen-specific performance of the test system). Medical judgment is required to interpret findings in light of correlative clinical information (e.g. age, gender, tumor stage, histologic and immunohistochemical findings), published literature, validation work, genotype/phenotype databases and other reliable sources. Importantly, the interpretation must address the medical question posed by the ordering physician, and also consider incidental findings of importance to the patient and their blood relatives.
The report submitted to the patient’s medical record documents findings and provides guidance to support clinical decision-making and follow-up. Such a report is best generated by a laboratory physician who is expert in disease pathobiology and in molecular technology, and who takes responsibility for the analyses and for the quality control processes supporting their interpretation.
In high complexity clinical laboratories in the United States (and in laboratories worldwide that are accredited by the College of American Pathologists or that meet equivalent standards), a laboratory physician must be available to discuss medical indications for testing and clinical implications of test results in a given patient. Published guidance outlines the principles of clinical grade genomic assay development, implementation and maintenance.119, 120, 121, 122, 123
Summary
This article describes recent advances in understanding gastric carcinogenesis. The findings imply that gastric adenocarcinoma is four separate diseases with respect to molecular pathogenesis. Prior epidemiologic studies may have overlooked the EBV molecular class that comprises only 9% of cancers, while emphasizing the chromosome instability class that comprises 50% of cancers. Furthermore, historic studies lumped cases with CpG island hypermethylation into a single group, but emerging evidence reveals the distinct patterns of hypermethylation in EBV-positive vs MSI molecular classes of cancer. Cancers of diffuse histology tend to be ‘genomically stable’ implying few mutations or gene amplifications, and prevalent defects in RHO signaling downstream of G-protein-coupled receptors.
Pathologists increasingly have the tools to support clinical trials of targeted therapy and to advance routine health care using ancillary molecular tests. The ‘GastroGenus Gastric Cancer Classifier’ is an example of a genomic test panel that adds value beyond histopathology of formalin-fixed cancer tissue by evaluating EBV status, MLH1 promoter methylation and multiple cancer gene sequences to interpret molecular class and to identify actionable mutations revealing clinical trial options or off-label use of an existing drug for metastatic cancer patients who fail standard therapy. On the horizon is integrative genomic technology combining gene expression profiles with DNA/RNA sequence information such as mutation, splice variants, fusion and pathogen identification. Advanced genomic technology shows promise to assist in early diagnosis, classification and monitoring of affected patients. Hopefully cancer prevention is also among the ways that genomic technology might improve public health.
References
Tang W, Morgan DR, Meyers MO, Dominguez RL, Martinez E, Kakudo K et al. Epstein–Barr virus infected gastric adenocarcinoma expresses latent and lytic viral transcripts and has a distinct human gene expression profile. Infect Agent Cancer 2012; 7: 21.
Hardbower DM, Peek RM Jr, Wilson KT . At the bench: Helicobacter pylori, dysregulated host responses, DNA damage, and gastric cancer. J Leukocyte Biol 2014; 96: 201–212.
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM . Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010; 127: 2893–2917.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D . Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90.
Kuo HY, Yeh KH . Molecular-targeted therapy for chemotherapy-refractory gastric cancer: a case report and literature review. Anticancer Res 2014; 34: 3695–3699.
Kasper S, Schuler M . Targeted therapies in gastroesophageal cancer. Eur J Cancer 2014; 50: 1247–1258.
Kim C, Mulder K, Spratlin J . How prognostic and predictive biomarkers are transforming our understanding and management of advanced gastric cancer. Oncologist 2014; 19: 1046–1055.
Fassan M, Simbolo M, Bria E, Mafficini A, Pilotto S, Capelli P et al. High-throughput mutation profiling identifies novel molecular dysregulation in high-grade intraepithelial neoplasia and early gastric cancers. Gastric Cancer 2014; 17: 442–449.
Wadhwa R, Song S, Lee JS, Yao Y, Wei Q, Ajani JA . Gastric cancer—molecular and clinical dimensions. Nat Rev Clin Oncol 2013; 10: 643–655.
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014; 513: 202–209.
Tsai KW, Liao YL, Wu CW, Hu LY, Li SC, Chan WC et al. Aberrant expression of miR-196a in gastric cancers and correlation with recurrence. Genes Chromosomes Cancer 2012; 51: 394–401.
Lo SS, Hung PS, Chen JH, Tu HF, Fang WL, Chen CY et al. Overexpression of miR-370 and downregulation of its novel target TGFbeta-RII contribute to the progression of gastric carcinoma. Oncogene 2012; 31: 226–237.
Oh HK, Tan AL, Das K, Ooi CH, Deng NT, Tan IB et al. Genomic loss of miR-486 regulates tumor progression and the OLFM4 antiapoptotic factor in gastric cancer. Clin Cancer Res 2011; 17: 2657–2667.
Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol 2010; 11: 136–146.
Suzuki H, Yamamoto E, Nojima M, Kai M, Yamano HO, Yoshikawa K et al. Methylation-associated silencing of microRNA-34b/c in gastric cancer and its involvement in an epigenetic field defect. Carcinogenesis 2010; 31: 2066–2073.
Yao Y, Suo AL, Li ZF, Liu LY, Tian T, Ni L et al. MicroRNA profiling of human gastric cancer. Mol Med Rep 2009; 2: 963–970.
Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D et al. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol 2009; 24: 652–657.
Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W et al. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest 2008; 88: 1358–1366.
Qu Y, Dang S, Hou P . Gene methylation in gastric cancer. Clin Chim Acta 2013; 424: 53–65.
Li L, Su X, Choi GC, Cao Y, Ambinder RF, Tao Q . Methylation profiling of Epstein–Barr virus immediate-early gene promoters, BZLF1 and BRLF1 in tumors of epithelial, NK- and B-cell origins. BMC Cancer 2012; 12: 125.
Zouridis H, Deng N, Ivanova T, Zhu Y, Wong B, Huang D et al. Methylation subtypes and large-scale epigenetic alterations in gastric cancer. Sci Transl Med 2012; 4: 156ra140.
Loh M, Liem N, Vaithilingam A, Lim PL, Sapari NS, Elahi E et al. DNA methylation subgroups and the CpG island methylator phenotype in gastric cancer: a comprehensive profiling approach. BMC Gastroenterol 2014; 14: 55.
Geddert H, Zur Hausen A, Gabbert HE, Sarbia M . EBV-infection in cardiac and non-cardiac gastric adenocarcinomas is associated with promoter methylation of p16, p14 and APC, but not hMLH1. Anal Cell Pathol (Amst) 2010; 33: 143–149.
Chang MS, Uozaki H, Chong JM, Ushiku T, Sakuma K, Ishikawa S et al. CpG island methylation status in gastric carcinoma with and without infection of Epstein–Barr virus. Clin Cancer Res 2006; 12: 2995–3002.
Yang P, Wang Y, Chen J, Li H, Kang L, Zhang Y et al. RCOR2 is a subunit of the LSD1 complex that regulates ESC property and substitutes for SOX2 in reprogramming somatic cells to pluripotency. Stem Cells 2011; 29: 791–801.
Suva ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 2014; 157: 580–594.
Zhao J, Liang Q, Cheung KF, Kang W, Lung RW, Tong JH et al. Genome-wide identification of EpsteinBarr virus-driven promoter methylation profiles of human genes in gastric cancer cells. Cancer 2013; 119: 304–312.
Liang Q, Yao X, Tang S, Zhang J, Yau TO, Li X et al. Integrative identification of Epstein–Barr virus-associated mutations and epigenetic alterations in gastric cancer. Gastroenterology (e-pub ahead of print 27 August 2014; doi:10.1053/j.gastro.2014.08.036.
Woellmer A, Hammerschmidt W . Epstein–Barr virus and host cell methylation: regulation of latency, replication and virus reactivation. Curr Opin Virol 2013; 3: 260–265.
Niller HH, Tarnai Z, Decsi G, Zsedenyi A, Banati F, Minarovits J . Role of epigenetics in EBV regulation and pathogenesis. Fut Microbiol 2014; 9: 747–756.
Okada T, Nakamura M, Nishikawa J, Sakai K, Zhang Y, Saito M et al. Identification of genes specifically methylated in Epstein–Barr virus-associated gastric carcinomas. Cancer Sci 2013; 104: 1309–1314.
Ryan JL, Jones RJ, Kenney SC, Rivenbark AG, Tang W, Knight ER et al. Epstein–Barr virus-specific methylation of human genes in gastric cancer cells. Infect Agent Cancer 2010; 5: 27.
Matsusaka K, Funata S, Fukayama M, Kaneda A . DNA methylation in gastric cancer, related to Helicobacter pylori and Epstein–Barr virus. World J Gastroenterol 2014; 20: 3916–3926.
Lin J, Gilbert J, Rudek MA, Zwiebel JA, Gore S, Jiemjit A et al. A phase I dose-finding study of 5-azacytidine in combination with sodium phenylbutyrate in patients with refractory solid tumors. Clin Cancer Res 2009; 15: 6241–6249.
Kenney SC, Mertz JE . Regulation of the latent-lytic switch in Epstein–Barr virus. Semin Cancer Biol 2014; 26: 60–68.
Tikhmyanova N, Schultz DC, Lee T, Salvino JM, Lieberman PM . Identification of a new class of small molecules that efficiently reactivate latent Epstein–Barr virus. ACS Chem Biol 2014; 9: 785–795.
Westphal EM, Blackstock W, Feng W, Israel B, Kenney SC . Activation of lytic Epstein–Barr virus (EBV) infection by radiation and sodium butyrate in vitro and in vivo: a potential method for treating EBV-positive malignancies. Cancer Res 2000; 60: 5781–5788.
Ghosh SK, Perrine SP, Williams RM, Faller DV . Histone deacetylase inhibitors are potent inducers of gene expression in latent EBV and sensitize lymphoma cells to nucleoside antiviral agents. Blood 2012; 119: 1008–1017.
Doolittle JM, Webster-Cyriaque J . Polymicrobial infection and bacterium-mediated epigenetic modification of DNA tumor viruses contribute to pathogenesis. MBio 2014; 5: e01015–14.
Perrine SP, Hermine O, Small T, Suarez F, O'Reilly R, Boulad F et al. A phase 1/2 trial of arginine butyrate and ganciclovir in patients with Epstein–Barr virus-associated lymphoid malignancies. Blood 2007; 109: 2571–2578.
Wildeman MA, Novalic Z, Verkuijlen SA, Juwana H, Huitema AD, Tan IB et al. Cytolytic virus activation therapy for Epstein–Barr virus-driven tumors. Clin Cancer Res 2012; 18: 5061–5070.
Gulley ML, Tang W . Using Epstein–Barr viral load assays to diagnose, monitor, and prevent posttransplant lymphoproliferative disorder. Clin Microbiol Rev 2010; 23: 350–366.
Heslop HE . Combining drugs and biologics to treat nasopharyngeal cancer. Mol Ther 2014; 22: 8–9.
Fukayama M, Hino R, Uozaki H . Epstein–Barr virus and gastric carcinoma: virus–host interactions leading to carcinoma. Cancer Sci 2008; 99: 1726–1733.
Ocean AJ, Christos P, Sparano JA, Shah MA, Yantiss RK, Cheng J et al. Phase II trial of bortezomib alone or in combination with irinotecan in patients with adenocarcinoma of the gastroesophageal junction or stomach. Invest New Drugs 2014; 32: 542–548.
Hui KF, Lam BH, Ho DN, Tsao SW, Chiang AK . Bortezomib and SAHA synergistically induce ROS-driven caspase-dependent apoptosis of nasopharyngeal carcinoma and block replication of Epstein–Barr virus. Mol Cancer Ther 2013; 12: 747–758.
Onoyama M, Kitadai Y, Tanaka Y, Yuge R, Shinagawa K, Tanaka S et al. Combining molecular targeted drugs to inhibit both cancer cells and activated stromal cells in gastric cancer. Neoplasia 2013; 15: 1391–1399.
Fu DX, Tanhehco Y, Chen J, Foss CA, Fox JJ, Chong JM et al. Bortezomib-induced enzyme-targeted radiation therapy in herpesvirus-associated tumors. Nat Med 2008; 14: 1118–1122.
Gressette M, Verillaud B, Jimenez-Pailhes AS, Lelievre H, Lo KW, Ferrand FR et al. Treatment of nasopharyngeal carcinoma cells with the histone-deacetylase inhibitor abexinostat: cooperative effects with cis-platin and radiotherapy on patient-derived xenografts. PLoS ONE 2014; 9: e91325.
Yang X, Dai W, Kwong DL, Szeto CY, Wong EH, Ng WT et al. Epigenetic markers for non-invasive early detection of nasopharyngeal carcinoma by methylation-sensitive high resolution melting. Int J Cancer (e-pub ahead of print 8 September 2014; doi:10.1002/ijc.29192).
Ahn SM, Chan JY, Zhang Z, Wang H, Khan Z, Bishop JA et al. Saliva and plasma quantitative polymerase chain reaction-based detection and surveillance of human papillomavirus-related head and neck cancer. JAMA Otolaryngol Head Neck Surg 2014; 140: 846–854.
Couraud S, Vaca-Paniagua F, Villar S, Oliver J, Schuster T, Blanche H et al. Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free dna in lung cancer from never-smokers: a proof-of-concept study from BioCAST/IFCT-1002. Clin Cancer Res 2014; 20: 4613–4624.
Li X, Zhou F, Jiang C, Wang Y, Lu Y, Yang F et al. Identification of a DNA methylome profile of esophageal squamous cell carcinoma and potential plasma epigenetic biomarkers for early diagnosis. PLoS ONE 2014; 9: e103162.
Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 2014; 158: 929–944.
Lee J, van Hummelen P, Go C, Palescandolo E, Jang J, Park HY et al. High-throughput mutation profiling identifies frequent somatic mutations in advanced gastric adenocarcinoma. PLoS ONE 2012; 7: e38892.
Tapia O, Riquelme I, Leal P, Sandoval A, Aedo S, Weber H et al. The PI3K/AKT/mTOR pathway is activated in gastric cancer with potential prognostic and predictive significance. Virchows Arch 2014; 465: 25–33.
Matsuoka T, Yashiro M . The role of PI3K/Akt/mTOR signaling in gastric carcinoma. Cancers (Basel) 2014; 6: 1441–1463.
Xing X, Zhang L, Wen X, Wang X, Cheng X, Du H et al. PP242 suppresses cell proliferation, metastasis, and angiogenesis of gastric cancer through inhibition of the PI3K/AKT/mTOR pathway. Anticancer Drugs 2014; 25: 1129–1140.
Zhang C, Awasthi N, Schwarz MA, Schwarz RE . The dual PI3K/mTOR inhibitor NVP-BEZ235 enhances nab-paclitaxel antitumor response in experimental gastric cancer. Int J Oncol 2013; 43: 1627–1635.
Wong CH, Loong HH, Hui CW, Lau CP, Hui EP, Ma BB et al. Preclinical evaluation of the PI3K-mTOR dual inhibitor PF-04691502 as a novel therapeutic drug in nasopharyngeal carcinoma. Invest New Drugs 2013; 31: 1399–1408.
Robles AI, Harris CC . Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harb Perspect Biol 2010; 2: a001016.
Fels Elliott DR, Fitzgerald RC . Molecular markers for Barrett’s esophagus and its progression to cancer. Curr Opin Gastroenterol 2013; 29: 437–445.
Shimizu T, Marusawa H, Matsumoto Y, Inuzuka T, Ikeda A, Fujii Y et al. Accumulation of somatic mutations in TP53 in gastric epithelium with Helicobacter pylori infection. Gastroenterology 2014; 147: 407–417 e3.
Lee JH, Kim SH, Han SH, An JS, Lee ES, Kim YS . Clinicopathological and molecular characteristics of Epstein–Barr virus-associated gastric carcinoma: a meta-analysis. J Gastroenterol Hepatol 2009; 24: 354–365.
Strong MJ, Xu G, Coco J, Baribault C, Vinay DS, Lacey MR et al. Differences in gastric carcinoma microenvironment stratify according to EBV infection intensity: implications for possible immune adjuvant therapy. PLoS Pathogen 2013; 9: e1003341.
NCCN National Comprehensive Cancer Network (NCCN) Clinical Practice Guidelines in Oncology: Gastric Cancer, version 1.2014. Available at http://www.nccn.org accessed 10 May 2014.
Liguigli W, Tomasello G, Toppo L, Ratti M, Passalacqua R . Ramucirumab for metastatic gastric or gastroesophageal junction cancer: results and implications of the REGARD trial. Fut Oncol 2014; 10: 1549–1557.
Lee J, Ou SH . Towards the goal of personalized medicine in gastric cancer—time to move beyond HER2 inhibition. Part II: Targeting gene mutations and gene amplifications and the angiogenesis pathway. Discov Med 2013; 16: 7–14.
Yamamoto H, Watanabe Y, Maehata T, Morita R, Yoshida Y, Oikawa R et al. An updated review of gastric cancer in the next-generation sequencing era: insights from bench to bedside and vice versa. World J Gastroenterol 2014; 20: 3927–3937.
Ott PA, Hodi FS, Robert C . CTLA-4 and PD-1/PD-L1 blockade: new immunotherapeutic modalities with durable clinical benefit in melanoma patients. Clin Cancer Res 2013; 19: 5300–5309.
Chen BJ, Chapuy B, Ouyang J, Sun HH, Roemer MG, Xu ML et al. PD-L1 expression is characteristic of a subset of aggressive B-cell lymphomas and virus-associated malignancies. Clin Cancer Res 2013; 19: 3462–3473.
Wang W, Sun J, Li F, Li R, Gu Y, Liu C et al. A frequent somatic mutation in CD274 3′-UTR leads to protein over-expression in gastric cancer by disrupting miR-570 binding. Hum Mutat 2012; 33: 480–484.
Yoda Y, Takeshima H, Niwa T, Kim JG, Ando T, Kushima R et al. Integrated analysis of cancer-related pathways affected by genetic and epigenetic alterations in gastric cancer. Gastric Cancer 2014.
Lee J, Ou SH . Towards the goal of personalized medicine in gastric cancer—time to move beyond HER2 inhibition. Part I: Targeting receptor tyrosine kinase gene amplification. Discov Med 2013; 15: 333–341.
Periasamy J, Muthuswami M, Rao DB, Tan P, Ganesan K . Stratification and delineation of gastric cancer signaling by in vitro transcription factor activity profiling and integrative genomics. Cell Signal 2014; 26: 880–894.
Nagarajan N, Bertrand D, Hillmer AM, Zang ZJ, Yao F, Jacques PE et al. Whole-genome reconstruction and mutational signatures in gastric cancer. Genome Biol 2012; 13: R115.
Lei Z, Tan IB, Das K, Deng N, Zouridis H, Pattison S et al. Identification of molecular subtypes of gastric cancer with different responses to PI3-kinase inhibitors and 5-fluorouracil. Gastroenterology 2013; 145: 554–565.
Thiem S, Pierce TP, Palmieri M, Putoczki TL, Buchert M, Preaudet A et al. mTORC1 inhibition restricts inflammation-associated gastrointestinal tumorigenesis in mice. J Clin Invest 2013; 123: 767–781.
Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet 2014; 46: 573–582.
Zhou J, Hayakawa Y, Wang TC, Bass AJ . RhoA mutations identified in diffuse gastric cancer. Cancer Cell 2014; 26: 9–11.
Kale VP, Hengst JA, Desai DH, Dick TE, Choe KN, Colledge AL et al. A novel selective multikinase inhibitor of ROCK and MRCK effectively blocks cancer cell migration and invasion. Cancer Lett 2014; 354: 299–310.
Tao J, Deng NT, Ramnarayanan K, Huang B, Oh HK, Leong SH et al. CD44-SLC1A2 gene fusions in gastric cancer. Sci Transl Med 2011; 3: 77ra30.
Lee J, Lee SE, Kang SY, Do IG, Lee S, Ha SY et al. Identification of ROS1 rearrangement in gastric adenocarcinoma. Cancer 2013; 119: 1627–1635.
Abel HJ, Al-Kateb H, Cottrell CE, Bredemeyer AJ, Pritchard CC, Grossmann AH et al. Detection of gene rearrangements in targeted clinical next-generation sequencing. J Mol Diagn 2014; 16: 405–417.
Yamamoto T, Iwatsuki K . Diversity of Epstein–Barr virus BamHI-A rightward transcripts and their expression patterns in lytic and latent infections. J Med Microbiol 2012; 61: 1445–1453.
Hino R, Uozaki H, Murakami N, Ushiku T, Shinozaki A, Ishikawa S et al. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res 2009; 69: 2766–2774.
Quinn LL, Zuo J, Abbott RJ, Shannon-Lowe C, Tierney RJ, Hislop AD et al. Cooperation between Epstein–Barr virus immune evasion proteins spreads protection from CD8+ T cell recognition across all three phases of the lytic cycle. PLoS Pathogen 2014; 10: e1004322.
Griffin BD, Gram AM, Mulder A, Van Leeuwen D, Claas FH, Wang F et al. EBV BILF1 evolved to downregulate cell surface display of a wide range of HLA class I molecules through their cytoplasmic tail. J Immunol 2013; 190: 1672–1684.
Lung RW, Tong JH, To KF . Emerging roles of small Epstein–Barr virus derived non-coding RNAs in epithelial malignancy. Int J Mol Sci 2013; 14: 17378–17409.
Marquitz AR, Mathur A, Chugh PE, Dittmer DP, Raab-Traub N . Expression profile of microRNAs in Epstein–Barr virus-infected AGS gastric carcinoma cells. J Virol 2014; 88: 1389–1393.
Marquitz AR, Mathur A, Shair KH, Raab-Traub N . Infection of Epstein–Barr virus in a gastric carcinoma cell line induces anchorage independence and global changes in gene expression. Proc Natl Acad Sci USA 2012; 109: 9593–9598.
Gulley ML, Tang W . Laboratory assays for Epstein–Barr virus-related disease. J Mol Diagn 2008; 10: 279–292.
Gulley ML, Pulitzer DR, Eagan PA, Schneider BG . Epstein–Barr virus infection is an early event in gastric carcinogenesis and is independent of bcl-2 expression and p53 accumulation. Hum Pathol 1996; 27: 20–27.
Kim L, Kim JM, Hur YS, Shin YW, Park IS, Choi SJ et al. Extended gastritis cystica profunda associated with Epstein–Barr virus-positive dysplasia and carcinoma with lymphoid stroma. Pathol Int 2012; 62: 351–355.
Schetter AJ, You WC, Lennette ET, Gail MT, Rabkin CS . Association of Epstein–Barr virus antibody levels with precancerous gastric lesions in a high-risk cohort. Cancer Sci 2008; 99: 350–354.
Zur Hausen A, van Rees BP, van Beek J, Craanen ME, Bloemena E, Offerhaus GJ et al. Epstein–Barr virus in gastric carcinomas and gastric stump carcinomas: a late event in gastric carcinogenesis. J Clin Pathol 2004; 57: 487–491.
Cheng N, Hui DY, Liu Y, Zhang NN, Jiang Y, Han J et al. Is gastric lymphoepithelioma-like carcinoma a special subtype of EBV-associated gastric carcinoma? New insight based on clinicopathological features and EBV genome polymorphisms. Gastric Cancer 2014.
Chen JN, Jiang Y, Li HG, Ding YG, Fan XJ, Xiao L et al. Epstein–Barr virus genome polymorphisms of Epstein–Barr virus-associated gastric carcinoma in gastric remnant carcinoma in Guangzhou, southern China, an endemic area of nasopharyngeal carcinoma. Virus Res 2011; 160: 191–199.
Tsai MH, Raykova A, Klinke O, Bernhardt K, Gartner K, Leung CS et al. Spontaneous lytic replication and epitheliotropism define an Epstein–Barr virus strain found in carcinomas. Cell Rep 2013; 5: 458–470.
Hutajulu SH, Hoebe EK, Verkuijlen SA, Fachiroh J, Hariwijanto B, Haryana SM et al. Conserved mutation of Epstein–Barr virus-encoded BamHI-A rightward frame-1 (BARF1) gene in Indonesian nasopharyngeal carcinoma. Infect Agent Cancer 2010; 5: 16.
Carneiro F, Seixas M, Sobrinho-Simoes M . New elements for an updated classification of the carcinomas of the stomach. Pathol Res Pract 1995; 191: 571–584.
Ryan JL, Morgan DR, Dominguez RL, Thorne LB, Elmore SH, Mino-Kenudson M et al. High levels of Epstein–Barr virus DNA in latently infected gastric adenocarcinoma. Lab Invest 2009; 89: 80–90.
Al-Batran SE, Werner D . Recent advances and future trends in the targeted therapy of metastatic gastric cancer. Expert Rev Gastroenterol Hepatol 2014; 8: 555–569.
Anonymous University of North Carolina Hospitals McLendon Clinical Laboratories. Available at https://labs.unchealthcare.org/directory/molecular_pathology accessed 10 May 2014.
Funkhouser WK Jr., Lubin IM, Monzon FA, Zehnbauer BA, Evans JP, Ogino S et al. Relevance, pathogenesis, and testing algorithm for mismatch repair-defective colorectal carcinomas: a report of the association for molecular pathology. J Mol Diagn 2012; 14: 91–103.
Pritchard CC, Smith C, Salipante SJ, Lee MK, Thornton AM, Nord AS et al. ColoSeq provides comprehensive lynch and polyposis syndrome mutational analysis using massively parallel sequencing. J Mol Diagn 2012; 14: 357–366.
Salipante SJ, Scroggins SM, Hampel HL, Turner EH, Pritchard CC . Microsatellite instability detection by next generation sequencing. Clin Chem 2014; 60: 1192–1199.
Perez-Cabornero L, Infante M, Velasco E, Lastra E, Miner C, Duran M . Evaluating the effect of unclassified variants identified in MMR genes using phenotypic features, bioinformatics prediction, and RNA assays. J Mol Diagn 2013; 15: 380–390.
McLean MH, El-Omar EM . Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol 2014; 11: 664–674.
Shi L, Campbell G, Jones WD, Campagne F, Wen Z, Walker SJ et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol 2010; 28: 827–838.
Tang W, Hu Z, Muallem H, Gulley ML . Quality assurance of RNA expression profiling in clinical laboratories. J Mol Diagn 2012; 14: 1–11.
Li S, Tighe SW, Nicolet CM, Grove D, Levy S, Farmerie W et al. Multi-platform assessment of transcriptome profiling using RNA-seq in the ABRF next-generation sequencing study. Nat Biotechnol 2014; 32: 915–925.
SEQC/MAQC-III Consortium. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat Biotechnol 2014; 32: 903–914.
Cottrell CE, Al-Kateb H, Bredemeyer AJ, Duncavage EJ, Spencer DH, Abel HJ et al. Validation of a next-generation sequencing assay for clinical molecular oncology. J Mol Diagn 2014; 16: 89–105.
Pritchard CC, Salipante SJ, Koehler K, Smith C, Scroggins S, Wood B et al. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J Mol Diagn 2014; 16: 56–67.
Cheng DT, Cheng J, Mitchell TN, Syed A, Zehir A, Mensah NY et al. Detection of mutations in myeloid malignancies through paired-sample analysis of microdroplet-PCR deep sequencing data. J Mol Diagn 2014; 16: 504–518.
Robb JA, Gulley ML, Fitzgibbons PL, Kennedy MF, Cosentino LM, Washington K et al. A call to standardize preanalytic data elements for biospecimens. Arch Pathol Lab Med 2014; 138: 526–537.
Korlimarla A, Prabhu JS, Anupama CE, Remacle J, Wahi K, Sridhar TS . Separate quality-control measures are necessary for estimation of RNA and methylated DNA from formalin-fixed, paraffin-embedded specimens by quantitative PCR. J Mol Diagn 2014; 16: 253–260.
Jennings L, Van Deerlin VM, Gulley ML . Recommended principles and practices for validating clinical molecular pathology tests. Arch Pathol Lab Med 2009; 133: 743–755.
Aziz N, Zhao Q, Bry L, Driscoll DK, Funke B, Gibson JS et al. College of American Pathologists' Laboratory Standards for next-generation sequencing clinical tests. Arch Pathol Lab Med 2014.
Gargis AS, Kalman L, Berry MW, Bick DP, Dimmock DP, Hambuch T et al. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol 2012; 30: 1033–1036.
Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, Brown KK, Deignan JL et al. ACMG clinical laboratory standards for next-generation sequencing. Genet Med 2013; 15: 733–747.
Schrijver I, Aziz N, Farkas DH, Furtado M, Gonzalez AF, Greiner TC et al. Opportunities and challenges associated with clinical diagnostic genome sequencing: a report of the Association for Molecular Pathology. J Mol Diagn 2012; 14: 525–540.
Acknowledgements
We thank colleagues at the University of North Carolina Department of Pathology and Laboratory Medicine, Lineberger Comprehensive Cancer Center and The Cancer Genome Atlas Network for supporting this work. This work was sponsored by the University of North Carolina (UNC) Department of Pathology and Laboratory Medicine, the University Cancer Research Fund, UNC Clinical Translational Science Award (NIH UL1 TR001111) and an award for Innovative Technologies for Molecular Analysis of Cancer (NCI R21 CA155543).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
MLG serves on an advisory board of Illumina and received assay validation reagents from Illumina.
Rights and permissions
This work is licensed under a Creative Commons Attribution 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/3.0/
About this article
Cite this article
Gulley, M. Genomic assays for Epstein–Barr virus-positive gastric adenocarcinoma. Exp Mol Med 47, e134 (2015). https://doi.org/10.1038/emm.2014.93
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/emm.2014.93
This article is cited by
-
Assessing Epstein–Barr virus in gastric cancer: clinicopathological features and prognostic implications
Infectious Agents and Cancer (2023)
-
Alteration in stemness causes exclusivity between Epstein–Barr virus-positivity and microsatellite instability status in gastric cancer
Gastric Cancer (2021)
-
PD-L1 expression combined with microsatellite instability/CD8+ tumor infiltrating lymphocytes as a useful prognostic biomarker in gastric cancer
Scientific Reports (2019)
-
Identification and characterization of an alternative cancer-derived PD-L1 splice variant
Cancer Immunology, Immunotherapy (2019)
-
Molecular biomarkers in gastro-esophageal cancer: recent developments, current trends and future directions
Cancer Cell International (2018)