Abstract
NM23 is a family of structurally and functionally conserved proteins known as nucleoside diphosphate kinases (NDPK). There is abundant mRNA expression of NM23-H1, NM23-H2, or a read through transcript (NM23-LV) in the primary sites of hepatocellular carcinoma (HCC). Although the NM23-H1 protein is implicated as a metastasis suppressor, the role of NM23-H2 appears to be less understood. Thus, the aim of this study was to examine whether NM23-H2 is associated with hepatocarcinogenesis. The level of NM23-H2 expression in tumor tissues and the surrounding matrix appeared to be independent of etiology and tumor differentiation. Its subcellular localization was confined to mainly the cytoplasm and to a lesser extent in the nucleus. Ectopic expression of NM23-H2 in NIH3T3 fibroblasts and HLK3 hepatocytes showed a transformed morphology, enhanced focus formation, and allowed anchorage-independent growth. Finally, NIH3T3 fibroblasts and HLK3 hepatocytes stably expressing NM23-H2 produced tumors in athymic mice and showed c-Myc over-expression. In addition, NF-κB and cyclin D1 expression were also increased by NM23-H2. Lentiviral delivery of NM23-H2 shRNA inhibited tumor growth of xenotransplanted tumors produced from HLK3 cells stably expressing NM23-H2. Collectively, these results indicate that NM23-H2 may be pro-oncogenic in hepatocarcinogenesis.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC) ranks fifth in frequency among all malignancies worldwide and causes approximately one million deaths annually (Schafer and Sorrell, 1999; Okuda, 2000). It has a heterogenous geographic distribution, with the greatest incidence in Asia and sub-Saharan Africa where hepatitis B is endemic (Bosch et al., 1999). The incidence of HCC has also been increasing steadily in the United States and Western Europe, owing to the increasing incidence of chronic hepatitis C (El-Serag and Mason, 1999). Cirrhosis, regardless of its etiology, is the main risk factor for HCC, but exposure to aflatoxin, alcohol, and/or an iron overload has also been associated with the development of HCC (Bosch et al., 1999). The etiology and carcinogenesis of HCC are multifactorial and multistage. The multistep process of HCC might involve chronic liver inflammation, cell death, cirrhosis and regeneration, DNA damage, dysplasia, and finally HCC (Tannapfel and Wittekind, 2002). Several genetic or epigenetic changes have been identified thus far. However, these alterations do not always reflect the biological or clinical characteristics of all HCCs.
The human NM23 family, which is characterized enzymatically as nucleoside diphosphate kinase (NDPK), consists of eight related genes that encode widely expressed proteins known as NM23-H1 through NM23-H8. NDPKs were originally identified as essential housekeeping enzymes that are required for the synthesis of nucleoside triphosphates by catalyzing the transfer of γ-phosphoryl groups from nucleoside triphosphates to nucleoside diphosphates and play a role in maintaining the intracellular nucleotide concentration (Lascu and Gonin, 2000). Altered NDPK expression has been reported to be involved in many cellular processes, including oncogenesis (Hailat et al., 1991), cellular proliferation (Keim et al., 1993), differentiation (Okabe-Kado et al., 1995), motility (Kantor et al., 1993), development (Dearolf et al., 1988), DNA repair (Postel et al., 2000), and apoptosis (Venturelli et al., 1995). Human NM23-H1 and NM23-H2 have 88% homology. Recently, NM23-LV, a read-through transcript from NM23-H1 into the neighboring NM23-H2 gene, has been reported (Valentijn et al., 2006). A variety of metastasis model systems have demonstrated that expression levels of NM23-H1, and to a lesser extent, NM23-H2, inversely correlate with metastatic potential (Rosengard et al., 1989; Bevilacqua, 1990; Nakayama et al, 1992; Steeg et al., 1993; Boissan et al., 2005). NM23-H1 and NM23-H2 are expressed abundantly in HCC (Yamaguchi et al., 1994). Accordingly, abundant NM23-H1 mRNA expression in the primary sites of a HCC is inversely correlated with the incidence of intrahepatic metastasis and the TNM stage. However, no correlation between the abundance of NM23-H2 mRNA and intrahepatic metastasis was reported (Yamaguchi et al., 1994).
NM23-H2 is a basic protein recently identified as the human PuF factor, which is a transcriptional activator of the c-Myc proto-oncogene (Postel et al., 1993; Dexheimer et al., 2009). Mutational analysis has identified residues and domains of NM23-H2 that are involved in DNA binding, suggesting a role in gene regulation (Postel et al., 2002). NM23-H2 was identified as a novel potential disease locus involved in mouse leukemic transformation (Joosten et al., 2002). Furthermore, NM23-H2 was reported to be significantly induced by 12-O-tetradecanoylphorbol-13-acetate (TPA) and UV radiation in both in vivo animal models and in vitro cell cultures, and NM23-H2 appears to play a key role in mediating the neoplastic transformation of epidermal cells in the early stages of skin carcinogenesis (Wei et al., 2004). However, the biological functions of NM23-H2 in oncogenesis are still unclear. Therefore, in order to understand the role of this protein in hepatocarcinogenesis, the biological properties of NM23-H2 in cellular transformation were characterized in vitro and in tumorigenicity in vivo.
Results
NM23-H2 expression in HCCs and hepatoma cell lines
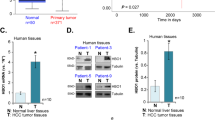
This aim of this study was to determine whether or not there is a correlation between NM23-H2 expression and the etiological cause or differentiation of tumor cells in HCC. The HCC samples examined in this study tested positive for the HBV serologic marker (58%). Using specific primer sets, real-time RT-PCR analysis was used to measure NM23-H1, NM23-H2, or NM23-LV mRNA levels in primary hepatocellular carcinomas as well as in normal and cirrhotic liver tissues (Figure 1A and Supplemental Figure S1A). NM23 mRNA was mostly over-expressed in primary HCCs compared with corresponding non tumor tissues (Figure 1B and Supplemental Figure S1B). According to Edmondson-Steiner's classification (Edmondson and Steiner, 1954), the level of NM23 mRNA induction was not significant between the better differentiated (n = 18) and the worse differentiated HCCs (n = 20). Furthermore, there was no significant difference in the level of NM23 mRNA induction between HBV-associated and non-HBV-associated HCCs. However, in HBV-associated HCCs, tumors with worse differentiation (GIII/IV) showed higher expression of NM23-H1 and -H2 than those that were better differentiated. Alternatively, we performed an unsupervised hierarchical clustering of gene expression profiles between non-tumors (20 normal livers and 20 cirrhotic livers) and HCC samples (40 from Jeonbuk National University Hospital and 60 from Seoul National University Hospital) using the Bead Chip DNA microarray method (Supplemental Figure S2). NM23-H1/2 mRNA was preferentially up-regulated in the HCC group. Next, we checked NM23-H2 protein expression and found that it was differentially over-expressed in 75% of the HCC tissues (9 out of 12) (Figure 1C). NM23-H2 protein expression in hepatoma cell lines compared with the non tumor cell line (THLE3) was also examined. NM23-H2 expression levels were high in HepG2, ALX, Huh7, SK-Hep-1, and SH-J1 hepatoma cells and weak in Hep3B and HLK2, and HLK3 cells (Figure 1D).
Increased NM23-H2 mRNA expression in HCC and hepatoma cell lines. (A) NM23-H1 and NM23-H2 each consist of five exons. NM23-LV contains the first four exons of NM23-H1 and last four exons of NM23-H2. Short bars indicate primers for real-time PCR. (B) The fold induction of NM23-H1, NM23-H2, or NM23-LV mRNA in individual HCCs (n = 38) and corresponding non tumor samples (seven normal liver and seven cirrhotic liver) were quantified and divided into two groups according to their association with differentiation (GI/II or GIII/IV, Edmondson-Steiner grading I/II or III/IV) or HBV. Real-time RT-PCRs were performed using three primer sets for NM23-H1, NM23-H2, or NM23-LV mRNA expression. Each bar represents the mean ± SE of three experiments. *P < 0.05; **P < 0.01; ***P < 0.0001. (C) NM23-H2 protein expression was determined in 12 HCC samples (T) and corresponding non tumor samples (N) according to tumor differentiation. Membranes were stripped and re-probed with actin antibody to confirm equal loading. (D) NM23-H2 protein expression in HCC, Chang liver, THLE3, HepG2, Hep3B, PLC/PRF/5 (Alexander), Huh7, SK-Hep-1, SH-J1, HLK2, and HLK3 cell lines. Membranes were stripped and reprobed with actin antibody to show equal loading.
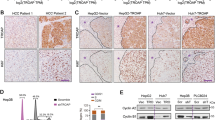
Immunoreactivity of NM23-H2 in HCC
We determined that the NM23-H2 antibody was specific for NM23-H2 immunogen in HEK293T cells transiently transfected with the NM23-H1 or NM23-H2 expression plasmid (Figure 2A). The NM23-H2 antibody (ATGP0490) used in this study did not show any cross reactivity with NM23-H1 or NM23-LV. Immunohistochemical staining for NM23-H2 revealed its over-expression was detected in various differentiations of HCC (Grade I to IV according to Edmondson-Steiner grading) and the surrounding tissues compared with non-tumor tissues (Figure 2B). Staining of NM23-H2 in HCC tissues was mainly cytoplasmic (Figure 2B, top middle and right panels), to a lesser extent nuclear (Figure 2B, bottom left panel), and in the surrounding fibrotic matrix (Figure 2B, bottom middle and right panels). In contrast, NM23-H1 was not observed in the surrounding fibrotic matrix (Figure 2C, right panel). The subcellular localization of NM23-H2 was examined by transiently transfecting cells with GFP fused to NM23-H2 and measuring the level of immunofluorescence (IF) simultaneously with a mouse monoclonal antibody (H2-206) used in previous studies (Tokunaga et al., 1993; Yamaguchi et al., 1994). IF with a rabbit polyclonal antibody (ATGP0490) yielded the same results. The expression of GFP fused to the NM23-H2 protein was confined mainly to the cytoplasm and to a lesser extent in the nuclei of HEK293T and HepG2 cells (Figure 2D), which corresponds to the immunoreactivity for the NM23-H2 protein. These results are consistent with a recent report (Bosnar et al., 2004).
NM23-H2 over-expression in HCC. (A) Specific immunoreactivity of NM23-H1 or NM23-H2 in cell lysates. Cells were transiently transfected with expression plasmids of NM23-H1 or NM23-H2. PC, parent cells; VC, vector control. (B) NM23-H2 over-expression in HCCs and surrounding matrix tissues according to Edmondson-Steiner grading (GI to GIII). (C) NM23-H1 was over-expressed in HCCs but not in surrounding matrix tissues. (D) Immunofluorescence assay showing ectopic expression is mainly localized in the cytoplasm and to a lesser extent in the nucleus of HEK293T and HepG2 cells. Hoechst 33258 staining of nuclei (blue). Immunoreactivity of NM23-H2 (red) is perfectively merged with GFP and NM23-H2 fusion proteins; bar, 20 µm.
Colony formation and cell transformation activities by NM23-H2
NM23-H2 transfected cells grown in culture had a transformed appearance, becoming irregularly elongated and losing their contact inhibition (Figure 3A). In order to determine if human NM23-H2 has oncogenic properties, the GFP-tagged NM23-H2 expression vector was used to generate cell lines to facilitate detection by fluorescence microscopy. Exogenous expression of NM23-H2 mRNA and expression of green fluorescence were confirmed in three independent NIH3T3 clones, NM1, NM5, and NM7 (Supplemental Figure S3A). The capacity of NH23-H2 to anchor independent cell growth was investigated by determining the level of cell growth as colonies in soft agar (Figures 3B and 3C), which was a property not observed in parental or vector control cells. Because occasional spontaneous transformations are known to occur in NIH3T3 cells, the capacity of NM23-H2 to enhance cell growth and transformation was further examined by measuring the ability of NM23-H2 to induce focus formation in Rat-1 fibroblasts (Bromberg et al., 1999). The ability to form foci in cell culture is a marker of cell transformation, which is believed to be a general property of an oncogene such as Ras (Land et al., 1983). The constitutively active and transforming Rasval12 allele was used as the positive control. The morphology of NM23-H2-induced foci was similar to that of Rasval12-induced foci (Supplemental Figure S3B).
Transformation ability of NM23-H2. (A) Transformed morphology of transfectants. Control NIH3T3 and HLK3 cells (left panels) and NM23-H2 transfectants (NM1, HNM10, and HNM14 cells, right panels) grown in culture for three days and observed using optical microscopy (original magnification, 200×). (B) NM23-H2-expressing NIH3T3 cells (NM1, NM5, and NM7) grow as colonies in soft agar, whereas vector control lines (VC1 and VC4) do not show any colonies. (C) NM23-H2-expressing HLK3 cells (HNM10 and HNM14) grow as colonies in soft agar, whereas vector control lines (HVC3 and HVC6) do not show any colonies. Colonies shown are 15 days old. The quantitative assay was performed in triplicate (lower). Each bar represents the means ± SE of threee experiments. **P < 0.01.
Xenotransplant and tumorigenicity
NIH3T3 cells stably expressing NM23-H2 (NM) formed tumor masses in mice (Figure 4A) compared with vector control (VC) or parental (NIH3T3) cells. Some transfectants (NM1 and NM5) formed bigger tumor masses than others (NM7). Tumor growth of NM23-H2 transfectants was significantly accelerated, but no tumor growth was observed for VC or NIH3T3 cells (Figure 4B). NM23-H2 immunoreactivity was mainly found in the cytoplasm and to a lesser extent in the nuclei of tumor tissues (Figure 4C). Stable transfectants expressing NM23-H2 showed c-Myc over-expression compared with vector control cells (Figure 4D). Immunohistochemistry showed that c-Myc over-expression was also detected in the nuclei of xenotransplanted tumor tissues (Figure 4E). Accordingly, NH23-H2-expressing HLK3 cells (HNM) were subcutaneously injected into nude mice in order to determine if NH23-H2 could enhance tumorigenicity. No tumor growth was observed in mice injected with vector control cells (HVC) (Figure 5A). Tumor growth was measured 58 days after inoculation (Figure 5B). Expression of NM23-H2 and hepatocyte antigen was observed by immunohistochemistry and confirmed by immunoblot analysis in xenotransplanted masses from HNM10 and HNM14 cells (Figure 5C, middle and bottom panels) as shown in human HCC samples (Figure 5C, top panels). In addition, the level of NM23-H2 appeared to induce in vivo tumorigenicity. NM23-H2 activates c-Myc transcription (Postel et al., 1993; Dexheimer et al., 2009). Therefore, we examined the level of c-Myc expression in cells cultured from xenografts and vector control cells. The level of c-Myc over-expression was at least two times higher in stable transfectants of HLK3 expressing NM23-H2 (HNM) and xenotransplanted tumor tissues positive for NM23-H2 (mHNM) compared with vector control cells (Figure 5D), which was confirmed by immunohistochemical staining of the nuclear or cytoplasmic fractions of the tumor tissues (data not shown). We also detected NF-κB and cyclin D1 expression in stable transfectants and xenotransplanted tumor tissues (mHNM). Next, an immunofluorescence assay revealed that transient expression of NM23-H2 co-localized with the expression of a subunit of c-Myc, cyclin D1, and NF-κB (p65) in HLK3 hepatocytes (Figure 5E). To provide support for the role of these factors in oncogenic regulation we analyzed the effects of NM23-H2 on the up-regulation of c-Myc, cyclin D1, and NF-κB in HLK3 cells after infection with adenoviral particles expressing NM23-H2. Transient expression of NM23-H2 up-regulated c-Myc, cyclin D1, and NF-κB in HLK3 cells (Supplemental Figure S4). These results suggest that c-Myc is not exclusively involved in NM23-H2-mediated hepatocarcinogenesis.
NM23-H2 expression enhances tumorigenicity. (A) NM23-H2-expressing NIH3T3 cells (NM1, NM5, and NM7) grow as tumors in nude mice, but there was no growth of parental (NIH3T3) or vector control (VC1 and VC4) cells. Tumors are shown 42 days post-injection. (B) Growth of tumor masses in NM23-H2-expressing (NM1, NM5, and NM7), parental (NIH3T3), and vector control (VC4) cells injected into nude mice. NM23-H2-expressing cell lines grew faster than control cells and formed tumor masses. Tumor volume was measured as a function of time (the animal was finally euthanized due to an ulceration of the primary tumor). Each value represents the mean ± SE. (C) Histology of NM23-H2-derived tumors stained with hematoxylin and eosin (upper panels). Expression of the NM23-H2 protein detected on independent immunohistochemistry experiments (lower panels). Immunoreactivity of NM23-H2 is visible in the nucleus and cytoplasm of anaplastic spindle cells (original magnification, top left and middle, and lower left panels, 100×; original magnification, top right, and bottom middle and right panels, 200×). (D) Western blot analysis showing c-Myc over-expression in xenotransplanted tumors inoculated with NIH3T3 cells stably expressing GFP-tagged NM23-H2 (NM1, NM5, and NM7). (E) Immunohistochemical staining of c-Myc in xenotransplanted tumors (top and bottom left panels) compared with negative staining with mouse IgG antibody (bottom left panel).
Oncogenic potential of NM23-H2 in HLK3 cells. (A) NM23-H2-expressing HLK3 cells (HNM10 and HNM14) but not vector control cells (HVC3 and HVC6) grow as tumors in nude mice. Tumors are shown 58 days post-injection. (B) Growth of the tumor mass in NM23-H2 expressing (HNM10 and HNM14) and vector control (HVC3 and HVC6) cells injected into nude mice. NM23-H2 expressing cell lines grew faster than control cells and formed tumor masses. The tumor volume was measured as a function of time (the animal was finally euthanized due to an ulceration of the primary tumor). Each value represents the mean ± SE. (C) Histology of NM23-H2-derived tumors stained with NM23-H2 (left panels) and hepatocyte (right panels) antibodies. HCC tissue staining was used as a positive control (original magnification, 200×). (D) Western blot analysis showing c-Myc, cyclin D1 (CCND1), and p65 over-expression in stable cell lines (HNM10 and HNM14), xenotransplanted tumors (mHNM10 and mHNM14), parent cells (HLK3), and vector control cells (HVC3 and HVC6). (E) Immunofluorescence assay showing that ectopic NM23-H2 expression overlaps with p65 (NF-κB), CCND1, and c-Myc over-expression in HLK3 cells. Hoechst 33258 staining of nuclei (blue). Immunoreactivity of NM23-H2 and Flag (FITC) is merged with c-Myc, CCND1, and p65 (TRITC); bar, 20 µm.
Knockdown of NM23-H2 led to inhibition of tumorigenicity
To further determine the functional role of NM23-H2 in HCC we generated a knockdown system of NM23-H2 by lentiviral delivery of shRNA into stable transfectants. We established knockdown cells barely expressing NM23-H2 from the stable cell lines, HNM10 and HNM14 (Figure 6A). Tumor growths were measured 30 days after xenotransplantation and compared with those of stable cells transduced with lentivirus containing non-target shRNA for NM23-H2. NM23-H2 silencing effectively inhibited tumor growth (Figure 6B). These results suggest that NM23-H2 silencing suppresses the pro-oncogenic function of NM23-H2.
Inhibition of tumorigenicity by knockdown of NM23-H2. (A) Immunoblot analysis was performed for NM23-H2 knockdown in NM23-H2 expressed cells (HNM10 and HNM14). Stable transfectants were transduced with lentivirus containing NM23-H2 or non-target (NT) shRNA. (B) Inhibition of tumor growth in stable transfectants transduced with lentivirus. The left and the right shoulders of mice were inoculated with non-targeting (shNT-HNM10 and shNT-HNM14) and targeting transfectants (sh221s-HNM10 and shNM465s-HNM14), respectively. Tumor growth was measured after 30 days (n = 6).
Discussion
The nm23-H1 gene was cloned on the basis of its potential as a metastasis suppressor gene (Steeg et al., 1988), whereas NM23-H2 was isolated based on sequence homology to NM23-H1 (Stahl et al., 1991). The earlier study revealed an inverse relationship between NM23-H1 expression and intrahepatic HCC metastases. However, there was no significant correlation between the expression of the NM23-H2 protein and clinicopathologic findings (Yamaguchi et al., 1994). The study by Yamaguchi et al. also showed that NM23-H2 mRNA is abundantly expressed in HCC tissues and established hepatoma cell lines, which correlated with neither the etiology nor the extent of tumor differentiation. Therefore, this study examined other aspects of oncogenic potential, such as the neoplastic transformation activity or the tumorigenicity of NM23-H2 in in vitro and in vivo systems.
Recently, the over-expression of mouse Nm23-M2 was reported to increase the level of foci formation and yield anchorage independent growth in mouse preneoplastic JB6 Cl 41-5a cells (Wei et al., 2004). Our study was also carried out using an expression vector containing human NM23-H2, and NIH3T3 and HLK3 cells were successfully transformed morphologically. The constitutive expression of the Ras protein family and other oncogenic proteins increases the focus-forming capability and decreases the level of growth contact inhibition of normal untransformed cells (Cox and Der, 1994). The present study revealed that ectopic expression of NM23-H2 increased the focus-forming capability and decreased the level of growth contact inhibition in Rat-1 cells, as shown in Rat-1 cells expressing Ras. GFP expression in foci and colonies was observed, and this GFP expression perfectively overlapped with the immunoreactivity for NM23-H2 in NIH3T3 cells. Furthermore, ectopic expression of NM23-H2 was correlated with an increased rate of growth in HLK3 and NIH3T3 cells. NM23-H2 not only accelerated tumorigenesis, but histological data also suggested that NM23-H2 promotes a characteristic microscopic phenotype. Accordingly, immortalized but non-tumorigenic breast MCF-10A cells progressing through the cell cycle show a low level of NME1 in the early phases (G0/G1), but subsequent peaking in the S-phase. In contrast, cells in the G0 phase express NME2, which peaks immediately following growth stimulation as cells enter the G1 phase (Caligo et al., 1995). In the case of immortalized fibroblasts transformed with SV40 large T antigen, expression of both NME1 and NME2 is increased compared to normal diploid cells (Ohneda et al., 1994). These observations suggest a role of NME members in the initial stages of tumorigenesis. Interestingly, murine NME2 (NM23-M2) is induced in response to chemical and UV radiation damage and has been shown to contribute to cancerous transformation of skin cells (Wei et al., 2004).
NM23-H2 has been reported to transactivate the c-Myc gene (Postel et al., 1993; Dexheimer et al., 2009), one of the most commonly activated oncogenes associated with the pathogenesis of liver tumors. Animal models have confirmed that Myc over-expression can induce HCC (Murakami et al., 1993; Wu et al., 2002). On the other hand, inhibition of Myc expression results in a loss of the neoplastic properties of a carcinoma (Simile et al., 2004). Recently, inactivation of the Myc oncogene has been reported to be sufficient for inducing sustained regression of invasive liver cancers (Shachaf et al., 2004). Therefore, targeted inactivation of the Myc oncogene may be an effective strategy for treating some liver cancers. Furthermore, Myc reactivation following its inactivation could immediately restore neoplastic properties. In the present study, the cells and tissues with NM23-H2 expression showed overt nuclear c-Myc over-expression. Therefore, it was found that NM23-H2 had pro-oncogenic potential both in vitro and in vivo. However, overexpression of c-Myc as well as other pro-oncogenic molecules such as cyclin D1 and NF-κB also appeared to be associated with hepatocarcinogenesis. Therefore, it will be important to further elucidate the molecular mechanism whereby NM23-H2 is involved in upstream hepatocarcinogenesis through cylcin D1 and NF-κB activation.
Recently, it was shown that NM23-H2 knockout mice have a normal phenotype at birth; however, these mice are defective for KCa3.1 ion channel activity and cytokine production from key immune cells (Di et al., 2010). Similarly, NM23-H1 knockout mice are also phenotypically normal but have reduced birth weight and show delayed mammary development (Arnaud-Dabernat et al., 2003). In comparison, double knockout of NM23-H1 and NM23-H2 results in undersized mice that die perinatally and show defects in hematopoiesis (Postel et al., 2009). NME2 has also been shown to be involved in non-cancerous pathological conditions such as immune disorders and atherosclerosis; murine models could provide valuable insights into the pathogenesis of these diseases (Srivastava et al., 2006; Rayner et al., 2007).
In conclusion, the tumorigenicity of NM23-H2 overexpressing cells depends on NM23-H2. Therefore, NM23-H2 with pro-oncogenic potential may be involved in hepatocarcinogenesis. Further understanding of the molecular mechanism underlying hepatocarcinogenesis and the targeted deregulation of this pro-oncogenic pathway might provide a strategy for effectively treating HCCs.
Methods
Tissue acquisition
Thirty-eight patients were selected and tested for either hepatitis B virus (HBV) or hepatitis C virus (HCV) markers in their sera using a Cobra Core EIA kit (F. Hoffmann-La Roche Ltd., Basel, Switzerland). Written informed consent was obtained from each patient. All 38 patients had HCC and underwent a curative liver resection. Paired samples of the tumor and corresponding cirrhotic non-tumor tissues were obtained from the resected liver specimens. Pathologists histologically confirmed and classified HCC according to Edmondson-Steiner's classification (Edmondson and Steiner, 1954). This protocol conformed to the ethical guidelines of the Institutional Review Board (IRB).
Cell culture and transfection
NIH3T3, HEK293T, Chang liver, Hep3B, PLC/PRF/5, HepG2, SK-HEP-1, Huh7, and Rat-1 cells were all obtained from the American Type Culture Collection (ATCC, Manassas, VA). Immortalized human hepatocytes (HLK3) and human HCC cells (HLK2 and SH-J1) were established and cultured in DMEM medium in our laboratory. In order to create a GFP/NM23-H2 fusion expression vector, the human NM23-H2 gene was PCR amplified using a forward primer containing the EcoRI restriction enzyme site (5'-CGGAATTCCCGGACC ATGGCCAAC-3') and a reverse primer containing the BamHI restriction enzyme site (5'-CGCGGAT CCGTCCACCTCTTATTC-3'). The PCR product of the open reading frame was ligated to pEGFP-C1 (BD Biosciences Clontech, Palo Alto, CA) at the EcoRI/BamHI site in frame. Transfections were performed using Lipofectamine reagent (Gibco-BRL, Grand Island, NY) according to the manufacturer's protocol. Stable transfectants were selected in the presence of G418 (600 µg/ml) over a two to three week period. Finally, the individual colonies were isolated using cloning rings. They were then expanded and assayed by Northern blot analysis to determine the expression level of the transfected gene. Cell proliferation was measured by trypan blue exclusion of cells plated in triplicate in a six-well plate. Rat-1 cells were similarly cultured and transfected with the pEJ6.6 plasmid containing activated H-rasVal-12, as described elsewhere (Bromberg et al., 1999).
Real-time RT-PCR
RNA was prepared from 100-200 mg of frozen tumor homogenized in 2 ml Trizol reagent (Invitrogen). Reverse transcription (RT) was performed using 2 µg of total RNA, 50 µM decamer, and 1 µl (200 units) of RT-PCR Superscript II (Invitrogen) at 37℃ for 50 min, as previously described. Specific primers for each gene were designed using the Primerdepot website (http://primerdepot.nci.nih.gov/) and are listed in Supplemental Figure 1A. The control 18S ribosomal RNA primer was purchased from Applied Biosystems (Foster City, CA) and used as the invariant control. The real-time RT-PCR reaction mixture consisted of 10 ng reverse-transcribed total RNA, 167 nM forward and reverses primers, and 2×PCR master mix in a final volume of 10 µl. PCR was performed in 384-well plates using the ABI Prism 7900HT Sequence Detection System (Applied Biosystems).
Animals and xenograft into nude mice
Eight-week-old female athymic nude mice (BALB/cByJ-Hfh11nu KRIBB, Daejeon, Korea) were used in all experiments. The animals were maintained in a specific pathogen-free environment. The animal room was kept at 20-22℃ under a 12-h light/dark cycle. Vector control cells or NM23-H2 transformed HLK3 and NIH3T3 cell lines (2×106) were resuspended in 0.2 ml of PBS and injected subcutaneously into the backs of the nude mice. Two perpendicular diameters of each tumor were measured using calipers, and the tumor volume was calculated using the method reported by the National Cancer Institute: length × width2 (in millimeters)/2 = volume (in cubic millimeters).
Soft agar assay
Trypsinized cells were resuspended in medium containing DMEM, 10% fetal bovine serum, antibiotics, and 3 ml of 0.3% noble agar (Difco, Sparks, MD). Cells (1×105 cells/well) were plated onto a solidified medium containing 5 ml of 0.7% noble agar in a 60 mm dish in six-well plates. The plates were incubated at 37℃ in 5% CO2, and fresh medium was added every four to five days. After two weeks, all colonies equal to or greater than 100 µm in diameter were counted.
Immunofluorescence, immunohistochemistry, and immunoblot analysis
NIH3T3, HepG2, and HEK293T cells grown on glass coverslips were fixed in 4% paraformaldehyde and permeabilized with 0.2% Triton X-100 in PBS. The coverslips were incubated for 1 h in PBS containing 0.1% BSA and a mixture of antibodies. After washing in PBS twice, the cells were incubated with tetramethylrhodamine isothiocyanate (TRITC) conjugated anti-rabbit antibody (1:100 dilution) (DAKO, Glostrup, Denmark) for 1 h. DNA was labeled using Hoechst dye 33258 (0.05 mg/ml). Laser scanning confocal microscopy was carried out using a Zeiss LSM510 with krypton-argon and helium-neon lasers, and a three-dimensional projection was generated using the accompanying software. Two types of antibodies specific for NM23-H2, a mouse monoclonal antibody (H2-206, Seikagaku, Tokyo, Japan) (Tokunaga et al., 1993; Yamaguchi et al, 1994) and a rabbit polyclonal antibody (ATGP0490, ATGen, Seongnam, Korea), were used for immunofluorescence, immunohistochemistry, and immunoblot analysis. Mouse monoclonal antibody to NM23-H1 (sc-465, Santa Cruz Biotechnology) or c-Myc (9E10, Santa Cruz Biotechnology) and rabbit polyclonal antibody against GFP (sc-8334, Santa Cruz Biotechnology) and mouse polyclonal antiboby against human IgG (Milipore, Billerica, MA) were used for immunoblot analysis.
Quantification and statistical analysis
Densitometric data was analyzed using the LAS-3000 system (Fuji Photo Film, Tokyo, Japan). Expression levels in each tumor tissue relative to non-tumor tissue were calculated by normalizing against the level of 18S ribosomal RNA. All data was entered into Microsoft Excel 5.0®, and GraphPad® Software was used to perform paired t-tests. P values < 0.05 were considered significant.
Abbreviations
- HCC:
-
hepatocellular carcinoma
- NDPK:
-
nucleoside diphosphate kinase
References
Arnaud-Dabernat S, Bourbon PM, Dierich A, Le MM, Daniel JY . Knockout mice as model systems for studying nm23/NDP kinase gene functions. Application to the nm23-M1 gene . J Bioenerg Biomembr 2003 ; 35 : 19 - 30
Bevilacqua G . NM23 gene expression and human breast cancer metastases . Pathol Biol (Paris) 1990 ; 38 : 774 - 775
Boissan M, Wendum D, Arnaud-Dabernat S, Munier A, Debray M, Lascu I, Daniel JY, Lacombe ML . Increased lung metastasis in transgenic NM23-Null/SV40 mice with hepatocellular carcinoma . J Natl Cancer Inst 2005 ; 97 : 836 - 845
Bosch FX, Ribes J, Borras J . Epidemiology of primary liver cancer . Semin Liver Dis 1999 ; 19 : 271 - 285
Bosnar MH, De Gunzburg J, Bago R, Brecevic L, Weber I, Pavelic J . Subcellular localization of A and B Nm23/NDPK subunits . Exp Cell Res 2004 ; 298 : 275 - 284
Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE . Stat3 as an oncogene . Cell 1999 ; 98 : 295 - 303
Caligo MA, Cipollini G, Fiore L, Calvo S, Basolo F, Collecchi P, Ciardiello F, Pepe S, Petrini M, Bevilacqua G . NM23 gene expression correlates with cell growth rate and S-phase . Int J Cancer 1995 ; 60 : 837 - 842
Cox AD, Der CJ . Biological assays for cellular transformation . Methods Enzymol 1994 ; 238 : 277 - 294
Dearolf CR, Tripoulas N, Biggs J, Shearn A . Molecular consequences of awdb3, a cell-autonomous lethal mutation of Drosophila induced by hybrid dysgenesis . Dev Biol 1988 ; 129 : 169 - 178
Dexheimer TS, Carey SS, Zuohe S, Gokhale VM, Hu X, Murata LB, Maes EM, Weichsel A, Sun D, Meuillet EJ, Montfort WR, Hurley LH . NM23-H2 may play an indirect role in transcriptional activation of c-myc gene expression but does not cleave the nuclease hypersensitive element III1 . Mol Cancer Ther 2009 ; 8 : 1363 - 1377
Di L, Srivastava S, Zhdanova O, Sun Y, Li Z, Skolnik EY . Nucleoside diphosphate kinase B knock-out mice have impaired activation of the K+ channel KCa3.1, resulting in defective T cell activation . J Biol Chem 2010 ; 285 : 38765 - 38771
Edmondson HA, Steiner PE . Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies . Cancer 1954 ; 7 : 462 - 503
El-Serag HB, Mason AC . Rising incidence of hepatocellular carcinoma in the United States . N Engl J Med 1999 ; 340 : 745 - 750
Hailat N, Keim DR, Melhem RF, Zhu XX, Eckerskorn C, Brodeur GM, Reynolds CP, Seeger RC, Lottspeich F, Strahler JR . High levels of p19/nm23 protein in neuroblastoma are associated with advanced stage disease and with N-myc gene amplification . J Clin Invest 1991 ; 88 : 341 - 345
Joosten M, Vankan-Berkhoudt Y, Tas M, Lunghi M, Jenniskens Y, Parganas E, Valk PJ, Löwenberg B, van den Akker E, Delwel R . Large-scale identification of novel potential disease loci in mouse leukemia applying an improved strategy for cloning common virus integration sites . Oncogene 2002 ; 21 : 7247 - 7255
Kantor JD, McCormick B, Steeg PS, Zetter BR . Inhibition of cell motility after nm23 transfection of human and murine tumor cells . Cancer Res 1993 ; 53 : 1971 - 1973
Keim DR, Hailat N, Kuick R, Reynolds CP, Brodeur GM, Seeger RC, Hanash SM . PCNA levels in neuroblastoma are increased in tumors with an amplified N-myc gene and in metastatic stage tumors . Clin Exp Metastasis 1993 ; 11 : 83 - 90
Land H, Parada LF, Weinberg RA . Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes . Nature 1983 ; 304 : 596 - 602
Lascu I, Gonin P . The catalytic mechanism of nucleoside diphosphate kinases . J Bioenerg Biomembr 2000 ; 32 : 237 - 246
Murakami H, Sanderson ND, Nagy P, Marino PA, Merlino G, Thorgeirsson SS . Transgenic mouse model for synergistic effects of nuclear oncogenes and growth factors in tumorigenesis: interaction of c-myc and transforming growth factor alpha in hepatic oncogenesis . Cancer Res 1993 ; 53 : 1719 - 1723
Nakayama T, Ohtsuru A, Nakao K, Shima M, Nakata K, Watanabe K, Ishii N, Kimura N, Nagataki S . Expression in human hepatocellular carcinoma of nucleoside diphosphate kinase, a homologue of the nm23 gene product . J Natl Cancer Inst 1992 ; 84 : 1349 - 1354
Ohneda K, Fukuda M, Shimada N, Ishikawa N, Ichou T, Kaji K, Toyota T, Kimura N . Increased expression of nucleoside diphosphate kinases/nm23 in human diploid fibroblasts transformed by SV40 large T antigen or 60Co irradiation . FEBS Lett 1994 ; 348 : 273 - 277
Okabe-Kado J, Kasukabe T, Hozumi M, Honma Y, Kimura N, Baba H, Urano T, Shiku H . A new function of Nm23/NDP kinase as a differentiation inhibitory factor, which does not require its kinase activity . FEBS Lett 1995 ; 363 : 311 - 315
Okuda K . Hepatocellular carcinoma . J Hepatol 2000 ; 32 : 225 - 237
Postel EH, Berberich SJ, Flint SJ, Ferrone CA . Human c-myc transcription factor PuF identified as nm23-H2 nucleoside diphosphate kinase, a candidate suppressor of tumor metastasis . Science 1993 ; 261 : 478 - 480
Postel EH, Abramczyk BM, Levit MN, Kyin S . Catalysis of DNA cleavage and nucleoside triphosphate synthesis by NM23-H2/NDP kinase share an active site that implies a DNA repair function . Proc Natl Acad Sci USA 2000 ; 97 : 14194 - 14199
Postel EH, Abramczyk BA, Gursky SK, Xu Y . Structure-based mutational and functional analysis identify human NM23-H2 as a multifunctional enzyme . Biochemistry 2002 ; 41 : 6330 - 6337
Postel EH, Wohlman I, Zou X, Juan T, Sun N, D'Agostin D, Cuellar M, Choi T, Notterman DA, La Perle KM . Targeted deletion of Nm23/nucleoside diphosphate kinase A and B reveals their requirement for definitive erythropoiesis in the mouse embryo . Dev Dyn 2009 ; 238 : 775 - 787
Rayner K, Chen YX, Hibbert B, White D, Miller H, Postel EH, O'Brien ER . NM23-H2, an estrogen receptor beta-associated protein, shows diminished expression with progression of atherosclerosis . Am J Physiol Regul Integr Comp Physiol 2007 ; 292 : R743 - R750
Rosengard AM, Krutzsch HC, Shearn A, Biggs JR, Barker E, Margulies IM, King CR, Liotta LA, Steeg PS . Reduced Nm23/Awd protein in tumour metastasis and aberrant Drosophila development . Nature 1989 ; 342 : 177 - 180
Schafer DF, Sorrell MF . Hepatocellular carcinoma . Lancet 1999 ; 353 : 1253 - 1257
Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, Yang Q, Bishop JM, Contag CH, Felsher DW . MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer . Nature 2004 ; 431 : 1112 - 1127
Simile MM, De Miglio MR, Muroni MR, Frau M, Asara G, Serra S, Muntoni MD, Seddaiu MA, Daino L, Feo F, Pascale RM . Down-regulation of c-myc and Cyclin D1 genes by antisense oligodeoxy nucleotides inhibits the expression of E2F1 and in vitro growth of HepG2 and Morris 5123 liver cancer cells . Carcinogenesis 2004 ; 25 : 333 - 341
Srivastava S, Li Z, Ko K, Choudhury P, Albaqumi M, Johnson AK, Yan Y, Backer JM, Unutmaz D, Coetzee WA, Skolnik EY . Histidine phosphorylation of the potassium channel KCa3.1 by nucleoside diphosphate kinase B is required for activation of KCa3.1 and CD4 T cells . Mol Cell 2006 ; 24 : 665 - 667
Stahl JA, Leone A, Rosengard AM, Porter L, King CR, Steeg PS . Identification of a second human nm23 gene, nm23-H2 . Cancer Res 1991 ; 51 : 445 - 449
Steeg PS, Bevilacqua G, Kopper L, irsson UP, Talmadge JE, Liotta LA, Sobel ME . Evidence for a novel gene associated with low tumor metastatic potential . J Nat Cancer Inst 1988 ; 80 : 200 - 204
Steeg PS, de la Rosa A, Flatow U, MacDonald NJ, Benedict M, Leone A . Nm23 and breast cancer metastasis . Breast Cancer Res Treat 1993 ; 25 : 175 - 187
Tannapfel A, Wittekind C . Genes involved in hepatocellular carcinoma: deregulation in cell cycling and apoptosis . Virchows Arch 2002 ; 440 : 345 - 352
Tokunaga Y, Urano T, Furukawa K, Kondo H, Kanematsu T, Shiku H . Reduced expression of nm23-H1, but not of nm23-H2, is concordant with the frequency of lymph-node metastasis of human breast cancer . Int J Cancer 1993 ; 55 : 66 - 71
Wei SJ, Trempus CS, Ali RC, Hansen LA, Tennant RW . 12-O-tetradecanoylphorbol-13-acetate and UV radiation-induced nucleoside diphosphate protein kinase B mediates neoplastic transformation of epidermal cells . J Biol Chem 2004 ; 279 : 5993 - 6004
Wu Y, Renard CA, Apiou F, Huerre M, Tiollais P, Dutrillaux B, Buendia MA . Recurrent allelic deletions at mouse chromosomes 4 and 14 in Myc-induced liver tumors . Oncogene 2002 ; 21 : 1518 - 1526
Yamaguchi A, Urano T, Goi T, Takeuchi K, Niimoto S, Nakagawara G, Furukawa K, Shiku H . Expression of human nm23-H1 and nm23-H2 proteins in hepatocellular carcinoma . Cancer 1994 ; 73 : 2280 - 2284
Valentijn LJ, Koster J, Versteeg R . Read-through transcript from NM23-H1 into the neighboring NM23-H2 gene encodes a novel protein, NM23-LV . Genomics 2006 ; 87 : 483 - 489
Venturelli D, Martinez R, Melotti P, Casella I, Peschle C, Cucco C, Spampinato G, Darzynkiewicz Z, Calabretta B . Overexpression of DR-nm23, a protein encoded by a member of the nm23 gene family, inhibits granulocyte differentiation and induces apoptosis in 32Dc13 myeloid cells . Proc Natl Acad Sci USA 1995 ; 92 : 7435 - 7439
Acknowledgements
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2011-0009814), the National R&D Program for Cancer Control (0620220), and the Korean Health Technology R&D Project (A101834), the Ministry of Health, Welfare and Family Affairs, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Experimental & Molecular Medicine website
Supplementary information
Rights and permissions
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Lee, MJ., Xu, DY., Li, H. et al. Pro-oncogenic potential of NM23-H2 in hepatocellular carcinoma. Exp Mol Med 44, 214–224 (2012). https://doi.org/10.3858/emm.2012.44.3.016
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3858/emm.2012.44.3.016
Keywords
This article is cited by
-
CTCF and EGR1 suppress breast cancer cell migration through transcriptional control of Nm23-H1
Scientific Reports (2021)
-
Implication of human endogenous retrovirus W family envelope in hepatocellular carcinoma promotes MEK/ERK-mediated metastatic invasiveness and doxorubicin resistance
Cell Death Discovery (2021)
-
Regulatory functions of Nm23-H2 in tumorigenesis: insights from biochemical to clinical perspectives
Naunyn-Schmiedeberg's Archives of Pharmacology (2015)
-
Insights into the biology and prevention of tumor metastasis provided by the Nm23 metastasis suppressor gene
Cancer and Metastasis Reviews (2012)