Abstract
TNF-α, a proinflammatory cytokine, inhibits osteoblast differentiation under diverse inflammatory conditions; however, the underlying mechanisms in terms of the TNF-α signaling pathway remain unclear. In this study, we examined the role of Msx2 in TNF-α-mediated inhibition of alkaline phosphatase (ALP) expression and the signaling pathways involved. TNF-α down-regulated ALP expression induced by bone morphogenetic protein 2 (BMP2) in C2C12 and Runx2-/- calvarial cells. Over-expression of Msx2 suppressed BMP2-induced ALP expression. Furthermore, TNF-α induced Msx2 expression, and the knockdown of Msx2 by small interfering RNAs rescued ALP expression, which was inhibited by TNF-α. TNF-α activated the NF-κB and the JNK pathways. Inhibition of NF-κB or JNK activation reduced the inhibitory effect of TNF-α on ALP expression, whereas TNF-α-induced Msx2 expression was only suppressed by the inhibition of the NF-κB pathway. Taken together, these results indicate that Msx2 mediates the inhibitory action of TNF-α on BMP2-regulated osteoblast differentiation and that the TNF-α-activated NF-κB pathway is responsible for Msx2 induction.
Similar content being viewed by others
Introduction
TNF-α is a proinflammatory cytokine that plays an important role in various physiological and pathological processes including cell death, growth, differentiation and inflammation (Chen and Goeddel, 2002; Wajant et al., 2003). In bone tissue, there is a long-standing interest in the role of TNF-α because of its association with osteoporosis and rheumatoid arthritis. TNF-α is overproduced in the bone marrow of postmenopausal osteoporosis patients and within the synovium of rheumatoid arthritis lesions, contributing to both systemic and local bone loss (Feldmann et al., 1996; Pfeilschifter et al., 2002). TNF-α increases bone resorption by promoting osteoclast differentiation and activation (Nanes, 2003; Boyce et al., 2005). In addition, TNF-α decreases osteoblastic bone formation through the promotion of osteoblast apoptosis, the inhibition of osteoblast differentiation and the suppression of the matrix protein expression such as osteocalcin by mature osteoblasts (Kuno et al., 1994; Kitajima et al., 1996; Nakase et al., 1997; Gilbert et al., 2000, 2002). To date, the most important mechanism of TNF-mediated inhibition of osteoblast differentiation is the regulation of Runx2, a critical transcription factor for osteoblast differentiation. TNF-α decreases both the mRNA and protein levels of Runx2 via the suppression of transcription, destabilization of mRNA, and enhanced degradation of Runx2 protein (Gilbert et al., 2002; Kaneki et al., 2006). Although the inhibitory roles of TNF-α in osteoblast differentiation have been described, the detailed TNF-α signaling pathways involved remain unclear.
Msx2 belongs to the homeobox transcription factor family, and Msx2 is involved in cranial bone development (Ferguson, 1994; Alappat et al., 2003). Msx2-deficient mice show defects in cranial bone formation, whereas transgenic mice over-expressing wild type Msx2 exhibit an overall increase in bone volume (Satokata et al., 2000; Cheng et al., 2008). In humans, haploinsufficiency of MSX2 causes parietal foramina; however, its gain-of-function mutation leads to craniosynostosis syndrome (Jabs et al., 1993; Wilkie et al., 2000). Despite these in vivo phenotypes, the exact function of Msx2 in osteoblast differentiation remains controversial. Several reports have shown that Msx2 promotes osteogenic differentiation but suppresses adipogenic differentiation (Cheng et al., 2003; Ichida et al., 2004). Alternatively, there have been several lines of opposing evidence showing that Msx2 suppresses the expression of bone marker genes, including Runx2, alkaline phosphatase (ALP), and osteocalcin (Shirakabe et al., 2001; Hassan et al., 2004; Kim et al., 2004). Thus, the role of Msx2 in osteoblast differentiation needs to be further clarified.
Recently, we showed that TNF-α promotes MSX2 expression in human vascular smooth muscle cells (Lee et al., 2010). Considering that TNF-α suppresses ALP activity (Gilbert et al., 2002) and that Msx2 inhibits ALP expression (Kim et al., 2004), we hypothesized that Msx2 may be a new target molecule that mediates the inhibitory action of TNF-α on osteoblast differentiation. Therefore, we examined the role of Msx2 in the TNF-α-mediated inhibition of ALP expression and the underlying regulatory mechanism in terms of the signal transduction pathway. In the present study, we showed that TNF-α induces Msx2 expression in C2C12 cells through NF-κB activation, which in turn inhibits bone morphogenetic protein 2 (BMP2)-induced expression of ALP.
Results
As an in vitro model system, C2C12, a murine mesenchymal precursor cell line, which can differentiate into several cell types such as myocytes, adipocytes, and osteoblasts, was used in this study (Lee et al., 2000). We first examined the effect of TNF-α on BMP2-induced osteoblast differentiation. C2C12 cells were osteogenically induced by BMP2 (100 ng/ml) treatment for 24 h. Osteogenic induction was verified by cytochemical staining of ALP, an early osteogenic marker. TNF-α inhibited BMP2-induced ALP activity in a dose-dependent manner (Figure 1A). TNF-α almost completely suppressed ALP activity at 10 ng/ml concentration. Consistent with ALP staining data, induction of ALP mRNA expression by BMP2 was also blocked by TNF-α (Figure 1B).
TNF-α suppresses BMP2-induced ALP expression in Runx2-/- cells. (A, B) C2C12 cells were incubated in DMEM supplemented with 5% FBS for 24 h in the presence of the indicated reagents. Runx2+/+ and Runx2-/- cells were incubated in α-MEM supplemented with 10% FBS, 10 mM β-glycerophosphate and 50 µg/ml ascorbic acid for 48 h in the presence of the indicated reagents. Then, ALP staining (A) or RT-PCR (B) was performed. (C) To block new protein synthesis, C2C12 cells were treated with BMP2 and/or TNF-α in the presence of cycloheximide (CHX, 10 µg/ml) for 24 h. The concentrations of TNF-α and BMP2 were 10 ng/ml and 100 ng/ml, respectively, or as otherwise indicated. *, In Runx2-/- cells, the RT-PCR products of the ALP gene were obtained by carrying out the amplification step for five more cycles than in C2C12 and Runx2+/+ cells.
Next, we examined the effect of TNF-α in Runx2-/- calvarial preosteoblast cells to analyze the involvement of target genes other than Runx2. Expectedly, BMP2 weakly induced ALP activity in Runx2-/- cells compared to C2C12 cells (Figure 1A). TNF-α also exerted an inhibitory effect on BMP2-induced ALP activity and mRNA expression in Runx2-/- cells (Figures 1A and 1B). The absence of Runx2 transcripts in Runx2-/- cells was confirmed by RT-PCR (Figure 1B). As a positive control, we used calvarial cells from wild type ICR mice. TNF-α also showed inhibitory effect on BMP2-induced ALP expression in Runx2+/+ cells but to a lesser degree compared to that in C2C12 cells and Runx2-/- cells (Figure 1B). Next, we observed the mRNA level of Smurf1, an E3 ubiquitin ligase, because it is involved in TNF-α-induced degradation of Runx2 and BMP signaling proteins (Kaneki et al., 2006; Guo et al., 2008). As previously reported, TNF-α stimulated the expression of Smurf1 in C2C12 and Runx2+/+ cells (Figure 1B); however, Smurf1 was not expressed in Runx2-/- cells, indicating that new target molecules other than Runx2 and Smurf1 may be involved in TNF-α-mediated inhibition of ALP activity in Runx2-/- cells.
To examine whether TNF-α-mediated inhibition of ALP expression requires new protein synthesis, we treated C2C12 cells with TNF-α and/or BMP2 in the presence of cycloheximide, a protein synthesis inhibitor, and then examined the expression of ALP mRNA. As shown in Figure 1C, TNF-α did not exert a significant inhibitory effect on ALP expression in the presence of cycloheximide, indicating that the suppressive activity of TNF-α on BMP2-induced ALP expression requires new protein synthesis.
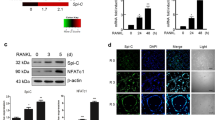
Before we examined the involvement of Msx2 in TNF-α-mediated ALP suppression, we first analyzed the effect of Msx2 on BMP2-induced ALP expression in our culture system. C2C12 cells were transiently transfected with pcDNA or an Msx2 expression vector and incubated with BMP2 for 24 h. As previously reported (Kim et al., 2004), BMP2-induced ALP expression was partially suppressed by Msx2 over-expression (Figure 2A). Next, we examined whether TNF-α regulates the expression of Msx2 in C2C12 cells. RT-PCR and immunoblot data demonstrated that TNF-α significantly up-regulated Msx2 expression at both the mRNA and protein level (Figure 2B, left and right panel); BMP2 alone did not increase Msx2 expression significantly; however, TNF-α-induced Msx2 expression was enhanced by BMP2. We also observed the effect of TNF-α on the expression of Msx2 in Runx2-/- cells. Similarly, TNF-α stimulated the expression of Msx2 in Runx2-/- cells as well as in Runx2+/+ cells (Figure 2C). Rescue of Runx2 expression in Runx2-/- cells did not change the Msx2 response to TNF-α (Figure 2D). Overall, these data suggest that Msx2 is a possible new TNF-α target molecule.
Expression of Msx2 is induced by TNF-α. (A) Over-expressed Msx2 inhibited BMP2-induced ALP expression. C2C12 cells were transiently transfected with pcDNA or the Msx2 expression vector and incubated in the presence of BMP2 for 24 h. RT-PCR was then performed. (B) C2C12 cells were incubated in the presence of the indicated reagents for 24 or 48 h. RT-PCR (left panel) or western blot analysis (right panel, nuclear extracts of 24 h samples) was performed to detect Msx2 induction. Histone H3 was used as a loading control. (C) Runx2+/+ and Runx2-/- cells were incubated in the presence of the indicated reagents for 24 h, and RT-PCR was performed. (D) Runx2 did not exert any effect on TNF-α-induced Msx2 expression. Runx2-/- cells were transiently transfected with pcDNA or the Runx2 expression vector and incubated in the presence of TNF-α for 24 h. RT-PCR was then performed.
To further define the role of Msx2 in TNF-α-inhibited ALP expression, we examined the expression levels of ALP after Msx2 knockdown by siRNA. The efficiency of Msx2 siRNA was verified by RT-PCR and immunoblot analysis (Figure 3). Msx2 siRNA clearly suppressed both basal and TNF-α-induced Msx2 expression. Furthermore, RT-PCR and ALP staining results showed that ALP mRNA expression and activity that were suppressed by TNF-α were partially rescued by Msx2 knockdown (Figure 3). These results indicate that TNF-α-induced Msx2 partially mediates the suppressive action of TNF-α on ALP expression.
TNF-α-inhibited ALP expression is rescued by the knockdown of Msx2. To induce the silencing of Msx2, C2C12 cells were transfected with siRNAs for Msx2 (si Msx2) or control siRNAs (si control) and further incubated in the presence of the indicated reagents for 24 h. Next, RT-PCR, immunoblot analysis and ALP staining were performed. Lamin B was used as a loading control.
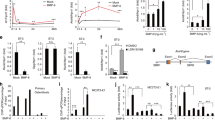
To define the link between the activation of the TNF receptor and Msx2 expression, we investigated TNF receptor downstream signaling pathways. TNF-α mainly activates MAPKs and the NF-κB pathway (Chen and Goeddel, 2002; Wajant et al., 2003). First, we examined the involvement of the NF-κB pathway in TNF activity. Immunoblot data showed that treatment with TNF-α increased IκBα phosphorylation within 10 min of TNF-α addition (Figure 4A, upper panel). BAY-11-7082, an NF-κB inhibitor, was used to block NF-κB activation. At the concentration used in this study (10 µM), BAY-11-7082 did not exhibit cytotoxicity or anti-proliferative activity (Figure 4B). RT-PCR and immunoblot data showed that BAY-11-7082 abrogated the induction of Msx2 expression by TNF-α (Figure 4A, middle panel). Consistent with the Msx2 results, TNF-α-inhibited ALP mRNA expression and activity were partially rescued by BAY-11-7082 treatment (Figure 4A, middle and lower panel). To rule out the nonspecific effects of BAY-11-7082, we over-expressed dnIκBα(S32A/S36A) in C2C12 cells to prevent NF-κB activation. These mutations render the protein non-phosphorylatable and consequently not degradable, inhibiting nuclear translocation of NF-κB (Brockman et al., 1995). Over-expression of dnIκBα effectively blocked TNF-α-induced nuclear translocation of RelA, a NF-κB p65 subunit (Figure 4C, upper panel). RT-PCR and immunoblot results showed that TNF-α-induced Msx2 expression was abolished when NF-κB was sequestered in the cytoplasm by dnIκBα (Figure 4C). Taken together, these results demonstrate that NF-κB activation is required for Msx2 induction by TNF-α.
TNF-α induces Msx2 expression via NF-κB activation. (A) Inhibition of NF-κB activation by BAY-11-7082 (BAY, 10 µM) blocked TNF-α-induced Msx2 expression. C2C12 cells were treated with the indicated reagents for 10 min (upper panel) or for 24 h (middle and lower panels), and then immunoblotting, RT-PCR and ALP staining were performed. The levels of phospho-IκBα and total IκBα were examined using whole cell lysates whereas the Msx2 protein level was examined using nuclear extracts. (B) BAY-11-7082 did not exert any cytotoxic effects at a concentration of 10 µM. The cell cytotoxicity/proliferation assay was performed using the CCK-8 reagent as described in Methods. Data represent the mean + SD of quadruplicates. *, P < 0.01 (Student's t-test) (C) Over-expression of dnIκBα suppressed TNF-α-induced Msx2 expression. C2C12 cells were transiently transfected with pcDNA or the dnIκBα expression vector and incubated in the presence of TNF-α for 24 h. Next, RT-PCR and immunoblot analyses were performed.
We also examined the effect of JNK activation on Msx2 expression because it has been reported that TNF-α inhibits BMP-induced osteoblast differentiation through JNK signaling (Mukai et al., 2007). TNF-α treatment increased the phosphorylation of JNK and its downstream target, c-Jun, within five min (Figure 5A). Although a previous report has shown that SP600125, a JNK inhibitor, promotes apoptosis in human leukemia cells (Moon et al., 2009), it did not exert cytotoxic effects on C2C12 cells at 10 µM concentration (Figure 5B). RT-PCR and immunoblot analyses revealed that Msx2 induction by TNF-α was not affected by SP600125 treatment. Apart from the effect on Msx2 expression, SP600125 partially rescued ALP mRNA expression that was suppressed by TNF-α (Figure 5C). These results suggest that the JNK pathway is not involved in TNF-α-induced Msx2 expression, but the pathway acts in TNF-α-mediated suppression of ALP expression.
JNK activation is not involved in TNF-α-induced Msx2 expression. (A, C) C2C12 cells were treated with the indicated reagents for 5 min (A) or for 24 h (C) and then immunoblot analysis and RT-PCR were performed. JNK activation was confirmed using whole cell lysates whereas the Msx2 protein level was examined using nuclear extracts. (B) SP600125 did not exert any cytotoxic effects at a concentration of 10 µM. Data represent the mean + SD of quadruplicates. *, P < 0.01 (Student's t-test)
Discussion
Previous reports have demonstrated that TNF-α inhibits spontaneous and BMP-induced osteoblast differentiation (Nakase et al., 1997; Gilbert et al., 2000). The major mechanistic findings of TNF-mediated osteoblast inhibition include suppression of Runx2 expression and diminution of BMP responses by reducing the levels of the BMP receptor and R-Smads (Gilbert et al., 2002; Kaneki et al., 2006; Singhatanadgit et al., 2006; Guo et al., 2008). Smurf1 promotes the proteasomal degradation of Runx2, the BMP receptor, and R-Smads by inducing ubiquitination. Smurf1 is also known to mediate TNF-induced systemic bone loss (Kaneki et al., 2006; Guo et al., 2008). In this study, based on the observations that TNF-α inhibited BMP2-induced ALP expression in Runx2-/- cells and that Smurf1 was not expressed in these cells, we deduced that molecules other than Runx2 and Smurf1 may also be involved in TNF-α-mediated suppression of osteoblast differentiation. Here, we present Msx2 as a novel target of TNF-α that mediates its inhibitory effect on ALP expression. We provide several lines of evidence implicating Msx2 in TNF-α-mediated suppression of osteoblast differentiation: (i) TNF-α induced the expression of Msx2 in both C2C12 cells and Runx2-/- cells; (ii) over-expression of Msx2 suppressed BMP2-induced ALP expression; (iii) knockdown of Msx2 expression by siRNA or blocking of Msx2 induction by the inhibition of NF-κB activation rescued ALP expression in the presence of TNF-α.
As described above, the biological function of Msx2 in osteoblast differentiation is currently controversial because studies have provided evidence of the role of Msx2 as both a negative or positive regulator (Komori, 2006). In this study, we aimed to define the role of Msx2 in the context of TNF-α-mediated inhibition of osteogenic differentiation. Our results showed that Msx2 plays a significant role in TNF-α-mediated inhibition of osteoblast differentiation, implicating Msx2 as a negative regulator of osteogenic differentiation. Molecular genetic studies have shown that Msx2 is essential for normal suture closure and skull mineralization (Jabs et al., 1993; Satokata et al., 2000; Wilkie et al., 2000; Cheng et al., 2008). Based on these in vivo phenotypes of Msx2 or MSX2 mutations, it has been thought that Msx2 acts as a positive regulator of osteoblast differentiation. A recent study, however, demonstrated that the Boston-type craniosynostosis MSX2 mutation (P148H) is not a gain-of-function mutation but is, instead, a loss-of-function mutation (Yoon et al., 2008). Yoon and colleagues showed that during murine calvarial development, Msx2 expression is localized predominantly in the suture mesenchyme and suggested that the primary function of MSX2 in calvarial bone development is to induce cell proliferation and suture maintenance but not to promote osteoblast differentiation (Yoon et al., 2008). In the cell culture system, the expression level of Msx2 in osteogenic lineage cells is regulated according to the stages of differentiation and maturation. For example, the expression level of Msx2 is maximum at the proliferating stage and subsequently decreases during the maturation of osteoblasts (Hassan et al., 2009). Moreover, Msx2 inhibits the function of key osteogenic regulators such as Runx2 and Dlx5 via protein-protein interactions and/or binding competition on DNA sequences, resulting in the suppression of the expression of the major osteogenic marker genes, ALP and osteocalcin (Zhang et al., 1997; Shirakabe et al., 2001; Hassan et al., 2004, 2006; Kim et al., 2004). In support of these reports, a recent study has also demonstrated that calvarial cells derived from Msx2-deficient mice are less proliferative and show a higher level of mineralization and bone differentiation marker gene expression than cells from wild type mice (Marijanović et al., 2009). In addition, it has been demonstrated that Msx2 acts as a molecular defense mechanism that prevents ossification of ligament and dermal fibroblasts (Yoshizawa et al., 2004; Hjelmeland et al., 2005). Taken together, these findings and our results indicate that Msx2 is a negative regulator during osteoblast differentiation.
To date, the detailed TNF-α signaling pathways involved in the inhibition of osteogenic differentiation remain unclear. Although a study has shown that TNF-α inhibits BMP-induced osteoblast differentiation through activation of JNK signaling (Mukai et al., 2007), no links were presented between JNK activation and a specific target gene. Here, we demonstrated that the activation of the NF-κB pathway is responsible for Msx2 induction triggered by TNF-α. These results were obtained by using both a chemical NF-κB inhibitor and dnIκBα over-expression. Previously, we have shown that cycloheximide does not affect TNF-α-induced MSX2 expression in human vascular smooth muscle cells (Lee et al., 2010), indicating that the signal cascades triggered by TNF-α directly induce Msx2 expression. Alternatively, the JNK/c-Jun pathway did not appear to be involved in the Msx2 induction by TNF-α because Msx2 expression was not affected by JNK inhibition. This finding does not rule out the involvement of the JNK/c-Jun pathway in TNF-α-mediated osteogenic inhibition because ALP expression suppressed by TNF-α was partially rescued by treatment with a JNK inhibitor. Our preliminary data suggest that Smurf1 appears to be the prime downstream target of JNK activation (data not shown), necessitating further studies.
In this study, BMP2 enhanced TNF-α-induced Msx2 expression even though there was a certain degree of variation in the magnitude. A previous report has shown that BMP2 rapidly induced the expression of homeodomain proteins such as Msx2 (2- to 3-fold) and Dlx5 (10- to14-fold) within 24 h in C2C12 cells (Hassan et al., 2006). In our study, however, BMP2 increased slightly Msx2 expression in some samples but not all (data not shown). It is not clear how BMP2 facilitated TNF-induced Msx2 expression but it is likely that in our culture system BMP2-activated signal was insufficient to induce Msx2 expression in itself but was able to heighten the Msx2 inducing signals by TNF. But further study is necessary to verify the signaling mechanism by which BMP2 enhance Msx2 expression.
In summary, our study reveals that the inhibitory action of TNF-α on BMP2-regulated osteoblast differentiation requires Msx2 as a mediator and that the TNF-α-activated NF-κB pathway is responsible for Msx2 induction. Our results support a functional role of Msx2 as a negative regulator of osteoblast differentiation.
Methods
Reagents and antibodies
Recombinant human TNF-α and BMP2 were purchased from R&D Systems (Minneapolis, MN). The easy-BLUE™ and StarTaq™ reagents were ordered from iNtRON Biotechnology (Sungnam, Korea) and the AccuPower RT-PreMix was purchased from Bioneer (Daejeon, Korea). PCR primers were synthesized by TaKaRa Korea (Seoul, Korea). The Cell Counting Kit-8 (CCK-8) was purchased from Dojindo Molecular Technologies (Gaithersburg, MA). BAY-11-7082 and SP600125 were from Calbiochem (Gibbstown, NJ). The Alkaline Phosphatase staining kit, cycloheximide, and anti-Msx2 antibody were purchased from Sigma (St. Louis, MO). Anti-NF-κB p65, anti-lamin B, anti-actin antibodies, and goat anti-rabbit HRP-conjugated IgG were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-IκBα, anti-phospho-IκBα, anti-JNK, anti-phospho-JNK, anti-c-Jun, anti-phospho-c-Jun, and anti-histone H3 antibodies were from Cell Signaling Technology (Danvers, MA). The Supex reagent for western blot analysis was ordered from Dyne-Bio (Sungnam, Korea). The NE-PER Nuclear and Cytoplasmic Extraction Reagent was obtained from PIERCE Biotechnology (Rockford, IL).
Cell culture, ALP staining, and transient transfection
C2C12 cells were cultured in DMEM supplemented with 10% FBS. To induce osteogenic differentiation, sub-confluent C2C12 cells were cultured in DMEM supplemented with 5% FBS and 100 ng/ml BMP2. Mouse calvarial osteoblasts (Runx2+/+ cells) were obtained from the frontal and parietal bones of neonatal ICR mice (Orient Bio Inc., Sungnam, Korea) as described previously (Kim et al., 2002). Cells were maintained in DMEM supplemented with 10% FBS. Runx2-/- cells, isolated from the primordium of calvarial tissue of Runx2-deficient mice (Lee et al., 2000), were maintained in α-MEM containing 10% FBS. For osteogenic differentiation, Runx2+/+ and Runx2-/- cells were cultured in α-MEM supplemented with 10% FBS, 100 ng/ml BMP2, 10 mM β-glycerophosphate and 50 µg/ml ascorbic acid.
To confirm the osteogenic differentiation, ALP staining was performed using the Alkaline Phosphatase staining kit according to the manufacturer's instructions.
For the transient transfection of expression vectors for Msx2, Runx2 and dominant negative (dn) IκBα, LipofectAMINE™ reagent was used.
RNA extraction and reverse transcription-PCR (RT-PCR)
To evaluate mRNA expression, semi-quantitative RT-PCR was performed in the range of linear amplification. Total RNA was isolated using easy-BLUE™ RNA Extraction Reagents. cDNA was synthesized from total RNA using the AccuPower™ RT-PreMix and subsequently subjected to PCR amplification using StarTaq™ polymerase. The PCR products were electrophoresed in a 1.2% agarose gel and visualized under UV light by ethidium bromide staining.
Mouse genes and their primer sequences for PCR were as follows (in parentheses, the annealing temperature used for the PCR reaction and the product size are described; f and r represent the forward and reverse primers, respectively): Msx2-f 5'-AACACAAGACCAACCGGAAG-3', Msx2-r 5'-GCCGTATATGGATGCTGCTT-3' (58℃, 299 bps); Runx2-f 5'-CCGCACGACAACCGCACCAT-3', Runx2-r 5'-CGCTCCGGCCCACAAATCTC-3' (56℃, 289 bps); Smad ubiquitin regulatory factor 1 (Smurf1)-f 5'-CTGGAGAACGAAGGAACAGT-3', Smurf1-r 5'-CGCAGTTCACTGTTAAGG-3' (56℃, 364 bps); ALP-f 5'-AGGCAGGATTGACCACGG-3', ALP-r 5'-TGTAGTTCTGCTCATGGA-3' (56℃, 439 bps); GAPDH-f 5'-TCACCATCTTCCAGGAGCG-3', GAPDH-r 5'-CTGCTTACCACCTTCTTGA-3' (56℃, 571 bps).
Cell cytotoxicity/proliferation assay
C2C12 cells were seeded at 1 × 103 cells/well in a 96-well plate. The following day, the adherent cells were fed with fresh differentiation medium containing 10 µM BAY-11-7082 or 10 µM SP600125. At the end of the indicated incubation periods, the cells were washed with medium and further incubated for 1 h with the CCK8 reagent mixture. The absorbance was then measured at 490 nm. The values represent the means of quadruplicate measurements.
Western blot analysis
After the appropriate treatments, whole cell lysates or nuclear extracts were prepared for western blot analysis. For the whole cell lysate preparation, cells were lysed in buffer consisting of 10 mM Tris-Cl (pH 7.5), 150 mM NaCl, 1 mM EDTA (pH 8.0), 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM sodium fluoride, 0.2 mM sodium orthovanadate, 1 mM PMSF, 1 µg/ml aprotinin, 1 µM leupeptin, and 1 µM pepstatin. The lysates were then sonicated briefly and centrifuged at 16,000 × g for 10 min, and the supernatants were used for immunoblot analysis. Nuclear extracts were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents according to the manufacturer's instructions. Protein concentrations were determined using a modified Bradford method. Each sample containing equal amounts of protein was subjected to SDS-PAGE. The proteins separated in the gel were subsequently transferred onto a PVDF membrane. The membrane was blocked with 5% nonfat dry milk in Tris-buffered saline containing 0.1% Tween20, incubated with the indicated primary antibody, and subsequently incubated with HRP-conjugated secondary antibody. Immune complexes were visualized using the Supex reagent and luminescence was detected with a LAS1000 (Fuji PhotoFilm; Tokyo, Japan).
Msx2 knockdown using small interfering (si) RNA
Msx2 siRNA and control siRNA (ON-TARGETplus Non-targeting siRNA #2 D-001210-02-05) were purchased from Dharmacon (Chicago, IL). Transfection into C2C12 cells was performed according to the manufacturer's instructions.
Abbreviations
- ALP:
-
alkaline phosphatase
- BMP2:
-
bone morphogenetic protein 2
- dnIκBα:
-
dominant-negative IκBα
- Smurf1:
-
Smad ubiquitin regulatory factor 1
References
Alappat S, Zhang ZY, Chen YP . Msx homeobox gene family and craniofacial development . Cell Res 2003 ; 13 : 429 - 442
Boyce BF, Li P, Yao Z, Zhang Q, Badell IR, Schwarz EM, O'Keefe RJ, Xing L . TNF-alpha and pathologic bone resorption . Keio J Med 2005 ; 54 : 127 - 131
Brockman JA, Scherer DC, McKinsey TA, Hall SM, Qi X, Lee WY, Ballard DW . Coupling of a signal response domain in I kappa B alpha to multiple pathways for NF-kappa B activation . Mol Cell Biol 1995 ; 15 : 2809 - 2818
Chen G, Goeddel DV . TNF-R1 signaling: a beautiful pathway . Science 2002 ; 296 : 1634 - 1635
Cheng SL, Shao JS, Charlton-Kachigian N, Loewy AP, Towler DA . MSX2 promotes osteogenesis and suppresses adipogenic differentiation of multipotent mesenchymal progenitors . J Biol Chem 2003 ; 278 : 45969 - 45977
Cheng SL, Shao JS, Cai J, Sierra OL, Towler DA . Msx2 exerts bone anabolism via canonical Wnt signaling . J Biol Chem 2008 ; 283 : 20505 - 20522
Feldmann M, Brennan FM, Maini RN . Rheumatoid arthritis . Cell 1996 ; 85 : 307 - 310
Ferguson MW . Craniofacial malformations: towards a molecular understanding . Nat Genet 1994 ; 6 : 329 - 330
Gilbert L, He X, Farmer P, Boden S, Kozlowski M, Rubin J, Nanes MS . Inhibition of osteoblast differentiation by tumor necrosis factor-alpha . Endocrinology 2000 ; 141 : 3956 - 3964
Gilbert L, He X, Farmer P, Rubin J, Drissi H, van Wijnen AJ, Lian JB, Stein GS, Nanes MS . Expression of the osteoblast differentiation factor RUNX2 (Cbfa1/AML3/Pebp2alpha A) is inhibited by tumor necrosis factor-alpha . J Biol Chem 2002 ; 277 : 2695 - 2701
Guo R, Yamashita M, Zhang Q, Zhou Q, Chen D, Reynolds DG, Awad HA, Yanoso L, Zhao L, Schwarz EM, Zhang YE, Boyce BF, Xing L . Ubiquitin ligase Smurf1 mediates tumor necrosis factor-induced systemic bone loss by promoting proteasomal degradation of bone morphogenetic signaling proteins . J Biol Chem 2008 ; 283 : 23084 - 23092
Hassan MQ, Javed A, Morasso MI, Karlin J, Montecino M, van Wijnen AJ, Stein GS, Stein JL, Lian JB . Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene . Mol Cell Biol 2004 ; 24 : 9248 - 9261
Hassan MQ, Tare RS, Lee SH, Mandeville M, Morasso MI, Javed A, van Wijnen AJ, Stein JL, Stein GS, Lian JB . BMP2 commitment to the osteogenic lineage involves activation of Runx2 by DLX3 and a homeodomain transcriptional network . J Biol Chem 2006 ; 281 : 40515 - 40526
Hassan MQ, Saini S, Gordon JA, van Wijnen AJ, Montecino M, Stein JL, Stein GS, Lian JB . Molecular switches involving homeodomain proteins, HOXA10 and RUNX2 regulate osteoblastogenesis . Cells Tissues Organs 2009 ; 189 : 122 - 125
Hjelmeland AB, Schilling SH, Guo X, Quarles D, Wang XF . Loss of Smad3-mediated negative regulation of Runx2 activity leads to an alteration in cell fate determination . Mol Cell Biol 2005 ; 25 : 9460 - 9468
Ichida F, Nishimura R, Hata K, Matsubara T, Ikeda F, Hisada K, Yatani H, Cao X, Komori T, Yamaguchi A, Yoneda T . Reciprocal roles of MSX2 in regulation of osteoblast and adipocyte differentiation . J Biol Chem 2004 ; 279 : 34015 - 34022
Jabs EW, Müller U, Li X, Ma L, Luo W, Haworth IS, Klisak I, Sparkes R, Warman ML, Mulliken JB . A mutation in the homeodomain of the human MSX2 gene in a family affected with autosomal dominant craniosynostosis . Cell 1993 ; 75 : 443 - 450
Kaneki H, Guo R, Chen D, Yao Z, Schwarz EM, Zhang YE, Boyce BF, Xing L . Tumor necrosis factor promotes Runx2 degradation through up-regulation of Smurf1 and Smurf2 in osteoblasts . J Biol Chem 2006 ; 281 : 4326 - 4333
Kim YH, Kim GS, Baek JH . Inhibitory action of bisphosphonates on bone resorption does not involve the regulation of RANKL and OPG expression . Exp Mol Med 2002 ; 34 : 145 - 151
Kim YJ, Lee MH, Wozney JM, Cho JY, Ryoo HM . Bone morphogenetic protein-2-induced alkaline phosphatase expression is stimulated by Dlx5 and repressed by Msx2 . J Biol Chem 2004 ; 279 : 50773 - 50780
Kitajima I, Soejima Y, Takasaki I, Beppu H, Tokioka T, Maruyama I . Ceramide-induced nuclear translocation of NF-kappa B is a potential mediator of the apoptotic response to TNF-alpha in murine clonal osteoblasts . Bone 1996 ; 19 : 263 - 270
Komori T . Regulation of osteoblast differentiation by transcription factors . J Cell Biochem 2006 ; 99 : 1233 - 1239
Kuno H, Kurian SM, Hendy GN, White J, deLuca HF, Evans CO, Nanes MS . Inhibition of 1,25-dihydroxyvitamin D3 stimulated osteocalcin gene transcription by tumor necrosis factor-alpha: structural determinants within the vitamin D response element . Endocrinology 1994 ; 134 : 2524 - 2531
Lee KS, Kim HJ, Li QL, Chi XZ, Ueta C, Komori T, Wozney JM, Kim EG, Choi JY, Ryoo HM, Bae SC . Runx2 is a common target of transforming growth factor beta1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12 . Mol Cell Biol 2000 ; 20 : 8783 - 8792
Lee HL, Woo KM, Ryoo HM, Baek JH . Tumor necrosis factor-alpha increases alkaline phosphatase expression in vascular smooth muscle cells via MSX2 induction . Biochem Biophys Res Commun 2010 ; 391 : 1087 - 1092
Marijanović I, Kronenberg MS, Ivkosić IE, Lichtler AC . Comparison of proliferation and differentiation of calvarial osteoblast cultures derived from Msx2 deficient and wild type mice . Coll Antropol 2009 ; 33 : 919 - 924
Moon DO, Kim MO, Kang CH, Lee JD, Choi YH, Kim GY . JNK inhibitor SP600125 promotes the formation of polymerized tubulin, leading to G2/M phase arrest, endoreduplication, and delayed apoptosis . Exp Mol Med 2009 ; 41 : 665 - 677
Mukai T, Otsuka F, Otani H, Yamashita M, Takasugi K, Inagaki K, Yamamura M, Makino H . TNF-alpha inhibits BMP-induced osteoblast differentiation through activating SAPK/JNK signaling . Biochem Biophys Res Commun 2007 ; 356 : 1004 - 1010
Nakase T, Takaoka K, Masuhara K, Shimizu K, Yoshikawa H, Ochi T . Interleukin-1 beta enhances and tumor necrosis factor-alpha inhibits bone morphogenetic protein-2-induced alkaline phosphatase activity in MC3T3-E1 osteoblastic cells . Bone 1997 ; 21 : 17 - 21
Nanes MS . Tumor necrosis factor-alpha: molecular and cellular mechanisms in skeletal pathology . Gene 2003 ; 321 : 1 - 15
Pfeilschifter J, Koditz R, Pfohl M, Schatz H . Changes in proinflammatory cytokine activity after menopause . Endocr Rev 2002 ; 23 : 90 - 119
Satokata I, Ma L, Ohshima H, Bei M, Woo I, Nishizawa K, Maeda T, Takano Y, Uchiyama M, Heaney S, Peters H, Tang Z, Maxson R, Maas R . Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation . Nat Genet 2000 ; 24 : 391 - 395
Shirakabe K, Terasawa K, Miyama K, Shibuya H, Nishida E . Regulation of the activity of the transcription factor Runx2 by two homeobox proteins, Msx2 and Dlx5 . Genes Cells 2001 ; 6 : 851 - 856
Singhatanadgit W, Salih V, Olsen I . Bone morphogenetic protein receptors and bone morphogenetic protein signaling are controlled by tumor necrosis factor-alpha in human bone cells . Int J Biochem Cell Biol 2006 ; 38 : 1794 - 1807
Wajant H, Pfizenmaier K, Scheurich P . Tumor necrosis factor signaling . Cell Death Differ 2003 ; 10 : 45 - 65
Wilkie AO, Tang Z, Elanko N, Walsh S, Twigg SR, Hurst JA, Wall SA, Chrzanowska KH, Maxson RE . Functional haploinsufficiency of the human homeobox gene MSX2 causes defects in skull ossification . Nat Genet 2000 ; 24 : 387 - 390
Yoon WJ, Cho YD, Cho KH, Woo KM, Baek JH, Cho JY, Kim GS, Ryoo HM . The Boston-type craniosynostosis mutation MSX2 (P148H) results in enhanced susceptibility of MSX2 to ubiquitin-dependent degradation . J Biol Chem 2008 ; 283 : 32751 - 32761
Yoshizawa T, Takizawa F, Iizawa F, Ishibashi O, Kawashima H, Matsuda A, Endo N, Kawashima H . Homeobox protein MSX2 acts as a molecular defense mechanism for preventing ossification in ligament fibroblasts . Mol Cell Biol 2004 ; 24 : 3460 - 3472
Zhang H, Hu G, Wang H, Sciavolino P, Iler N, Shen MM, Abate-Shen C . Heterodimerization of Msx and Dlx homeoproteins results in functional antagonism . Mol Cell Biol 1997 ; 17 : 2920 - 2932
Acknowledgements
This work was supported by the Korea Science and Engineering Foundation Grant M10646010002-06N4601-00210 funded by the Korean government (MOST) and by the Korea Research Foundation Grant funded by the Korean Government (MOEHRD, Basic Research Promotion Fund) (KRF-2006-000-E00093, KRF-2007-000-E00073).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Lee, HL., Yi, T., Woo, K. et al. Msx2 mediates the inhibitory action of TNF-α on osteoblast differentiation. Exp Mol Med 42, 437–445 (2010). https://doi.org/10.3858/emm.2010.42.6.045
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3858/emm.2010.42.6.045
Keywords
This article is cited by
-
Expression of VEGFA-mRNA in classical and MSX2-mRNA in non-classical monocytes in patients with spondyloarthritis is associated with peripheral arthritis
Scientific Reports (2021)
-
KLF4 as a rheostat of osteolysis and osteogenesis in prostate tumors in the bone
Oncogene (2019)
-
Short-Term Effects of TNF Inhibitors on Bone Turnover Markers and Bone Mineral Density in Rheumatoid Arthritis
Calcified Tissue International (2016)
-
Nonalcoholic fatty liver disease and decreased bone mineral density: is there a link?
Journal of Endocrinological Investigation (2015)
-
Sirt1 overexpression protects murine osteoblasts against TNF-α-induced injury in vitro by suppressing the NF-κB signaling pathway
Acta Pharmacologica Sinica (2012)