Abstract
Transforming growth factor-beta (TGF-β) and its receptors have been suggested to play key roles in the pathogenesis of asthma. The aim of this study was to evaluate the effects of genetic variations in the TGF-β receptor type III (TGFBR3) on asthma and on its related phenotypes in the general population. A cohort of 2,118 subjects aged from 10 to 18 years responded to a questionnaire concerning asthma symptoms and risk factors. Methacholine airway hyperresponsiveness (AHR), skin test responses to common aeroallergens, and serum total IgE levels were evaluated in the cohort. A total of 19 SNPs for TGFBR3 were found using direct re-sequencing in 24 healthy adults. Of these, informative SNPs [+44T>C (S15F) and +2753G>A at 3'UTR] were selected and scored using the high throughput single base extension method. Atopy was identified in subjects with 44T>C allele [P = 0.04, OR (95% CI) = 0.79 (0.62-0.99)] and in subjects with Ht1 (CG) more frequently than in subjects with other haplotypes [P = 0.04, OR (95% CI) = 1.27 (1.01-1.59)]. The A allele in 2753G>A was more common in subjects with non-atopic asthma [OR (95% CI) = 1.76 (1.01-3.05)]. A significant association was found between non-atopic asthma and 44T_2753A [OR (95% CI) = 2.16 (1.22-3.82)]. Genetic variations in TGFBR3 appear to be associated with a genetic predisposition to development of asthma and to phenotypes of asthma. Also, the minor allele 2753G and the haplotype TA in the TGFBR3 gene were associated with a pathogenesis of non-atopic asthma.
Similar content being viewed by others
Introduction
Asthma is a chronic inflammatory disorder of the airways that is characterized by airway hyperresponsiveness (AHR) and airway remodeling (Holgate, 2002). Asthma is a complex phenotype that is clinically difficult to define. This difficulty is associated with intermediate phenotypes, such as atopy and AHR, that have provided useful objective alternatives in genetic and epidemiologic studies (Wiesch et al., 1999; Kim et al., 2002; Townley and Horiba, 2003).
Transforming growth factor beta (TGF-β) is a secreted protein that regulates proliferation, differentiation, and death in various cell types (Moustakas et al., 2002). In addition to impacting embryogenesis, cancer, and matrix formation, it has profound effects on the immune system (Chen and Wahl, 2002). Deletion of TGF-β in mice leads to spontaneous T cell activation, infiltration of lymphocytes and macrophages into multiple organs, and massive inflammation leading to death in mice 3-4 weeks of age (Shull et al., 1992). TGF-β appears to have both pro- and anti-inflammatory functions and may participate in the initiation, progression, and resolution of inflammatory and immune responses in the airways (Catherine et al., 2003; Carsten et al., 2004). TGF-β is also involved in airway remodeling in asthma, being implicated in connective tissue remodeling, repair, and fibrosis (Mauviel, 2005; Wiebke et al., 2005).
In TGF-β signaling pathways, three TFG-β receptors (TGFBRs) have been identified, including type I, II, and type III receptors. Upon TGF-β binding to type II receptors, TGF-β RI is subsequently recruited and phosphorylated. TGF-β initiates cellular signaling by either binding to type III receptors, which then present TGF-β to the type II receptors, or by binding to type II receptors directly (Blobe et al., 2001). However, common signal transduction is performed through a heteromeric complex of type I and II transmembrane serine/threonine kinase receptors (Dijke and Hill, 2004). Although TGF-β type III receptors are thought to have an accessory function, they have an essential role in facilitating TGF-β binding (Sankar et al., 1995).
There are several reports of association studies for TGF-β gene polymorphism and asthma (Nagpal et al., 2005; Mak et al., 2006). Nagpal et al. showed an association between -800G/A and -509C/T polymorphisms in the TGF-β gene in an Indian population with asthma, and -509C/T also affected the serum levels of TGF-β1. TGF-β1 gene polymorphism was identified in Chinese patients with asthma in Hong Kong, and -509C/T and 869T/C were associated with asthma susceptibility in atopic subjects. SNP may play a role in the pathogenesis of airflow obstruction. This needs references Research involving TGF-β receptors is, so far, lacking. TGF-β receptors have an important function in the signal transduction process in mediation of the biological effects of cytokine. This study focused on TGFBR3 and we evaluated genetic variations associated with asthma and its intermediate phenotypes atopy and AHR in a cohort of 2,118 children and adolescents in a Korean population.
Results
Screening and scoring of SNPs in the TGFBR3 gene
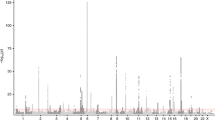
A total of 19 SNPs of TGFBR3 were screened to identify informative SNPs (minor allele frequency >2%) (Table 1). Two informative SNPs in the TGFBR3 gene, +44C>T [S15F (Serine → Phenylalanine at codon 15)] in exon 3 and 2753G>A (3'UTR)-exon 18, were identified (Figure 1). The distributions of these two loci were in a Hardy - Weinberg equilibrium (P = 0.12 for S15F and P = 0.56 for 2753G>A). The genotype frequencies of individual SNPs in the general population were examined and four haplotypes [Ht1 (CG), Ht2 (TG), Ht3 (TA), and Ht4 (CA)] were reconstructed (Table 2).
Genomic organization of TGFBR III and pairwise linkage disequilibium of 19 informative SNPs in the general population. The SNPs selected for scoring are shown in bold. Individual identified SNPs and their locations are shown in the gene. Distributions of 19 screened SNPs of TGFBR3 (minor allele frequency>2%) on human chromosome 1p33-p32. Bold letters indicate locations of the informative SNPs, +44C>T [S15F (Serine → Phenylalanine at codon 15)] in exon 3 and 2753G>A (3'UTR) in exon 18 that were identified.
Genetic effects of individual SNPs in the TGFBR3 gene on IgE responses to allergens
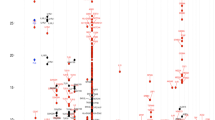
Th2 sensitization to aeroallergens is an important risk factor for asthma and AHR and is known to have genetic components. Thus, we evaluated the association of genetic variations in the TGFBR3 gene with IgE responses to common aeroallergens in the general population. Atopy (defined as a positive skin test response to one or more common aeroallergens) was significantly associated with 44T>C allele [P = 0.04, OR (95% CI) = 0.79 (0.62-0.99) in a recessive model of the minor allele]; however, this phenotype was not associated with +2753G>A polymorphism (Table 2). Haplotype analysis showed that atopy was more common in subjects with Ht1 (CG) than in subjects with other haplotypes [P = 0.04, OR (95% CI) = 1.27 (1.01-1.59) respectively, in a dominant model] (Figure 2).
Association between the haplotypes of the skin index and total IgE (Ht1 dominant model). Haplotype analysis showed that atopy was greater in subjects with Ht1 (CG) than in subjects with the other haplotypes [P = 0.04, OR (95% CI) = 1.27 (1.01-1.59) in a dominant model]. Haplotype analysis of total serum IgE showed that Ht1 was more prevalent than other haplotypes (P = 0.008 in a dominant model).
Effects of individual SNPs and their haplotypes in the TGFBR3 gene on asthma according to atopic status
We evaluated associations between variations in the TGFBR3 gene and asthma according to atopic status in the general population. The prevalence of asthma (defined as a current wheezing plus a positive AHR) was significantly greater in subjects with the +2753A [P = 0.01, OR (95% CI) = 1.66 (1.12-2.45) in a dominant model of the minor allele] (Tables 4 and 5).
Haplotype analysis showed that the asthma prevalence was significantly greater in subjects with Ht3 (TA) versus other haplotypes [P = 0.01, OR (95% CI) = 1.72 (1.12-2.64) in a dominant model. In terms of the genetic effects of SNPs of the TGFBR3 gene on atopic and non-atopic asthma, among atopic subjects the frequencies of individual SNPs of S15F and +2753G>A were similar in asthmatic subjects. The S15F and +2753G>A haplotypes were similar in atopic asthmatic subjects. However, among non-atopic subjects, the A allele in +2753G>A was significantly more common in subjects with asthma than in non-asthmatic subjects (P = 0.04 in a dominant model of the minor allele), although S15F polymorphism was not associated with asthma prevalence. Haplotype analysis showed that, among non-atopic subjects, the asthma prevalence was significantly greater in subjects with Ht3 (TA) [P = 0.007, OR (95% CI) = 2.16 (1.22-3.82) in a dominant model] (Table 6).
Discussion
The TGFBR3 gene is located on 1p33-p32 and encodes a protein (300 kDa) of 849 amino acids with a single transmembrane domain and a short stretch of the intracellular domain (Morén et al., 1992). We screened 19 SNPs of TGFBR3 using direct re-sequencing, 13 of which (68%) were available on a public database. Two informative SNPs, +44C>T (S15F) and 2753G>A (3'UTR), were selected for genetic consideration.
The number of examined individuals or families is important when drawing conclusions from genetic association studies. Greater statistical power can be achieved by increasing the sample size than by increasing the number of polymorphisms, and the sampling of 500 individuals provided sufficient repeatability to detect the presence of causative polymorphisms with a relatively small effect (Long and Langley, 1999). Thus, the more than 2,000 subjects involved in this study provided for strong repeatability.
SNPs are promising genetic markers for the purpose of complex disease gene hunting. Almost 5 million of the estimated 10 million SNPs have been identified to date and, of these, over 3 million have been assigned as "rsSNPs" in dbSNP (NCBI dbSNP build 110). However, it has been demonstrated that these genetic variations show significant ethnic differences. Of the 458 SNPs in 161 disease candidate genes collected from a publicly available SNP database, 43.9% were polymorphic in the Korean population, whereas 44.5% were monomorphic (Han et al., 2004).
Association studies of cSNPs within candidate genes, which result in amino acid changes, have the potential to identify genetic factors in complex genetic disorders, including asthma, and are generally more powerful than linkage studies in this respect (Risch and Merikangas, 1996). Thus, we tagged SNPs in the TGFBR3 gene and found two informative SNPs.
Genetic variations of the T allele in 44C>T SNP of TGFBR3 appeared to be associated with reduced IgE responses to common environmental aeroallergens. However, atopy was more prevalent in subjects with Ht1 (CG) haplotypes. It is likely that S15F in TGFBR3 is a more important genetic marker for predicting Th2 sensitization to an allergen than is +2753G>A.
Airway allergic inflammation and AHR are the hallmarks of asthma and appear to be central to the pathogenesis of asthma. With respect to the immunological pathogenesis of asthma, Th2 cytokines, such as IL-4, IL-5, IL-9, and IL-13, are considered to be key mediators of the development of asthma phenotypes, such as airway inflammation and AHR (Mosmann et al., 1986; Kim et al., 2008). IL-13 is now believed to be a central mediator in the development of allergic asthma (Wills-Karp et al., 1998). TGF-β1 is a key downstream mediator in the development of IL-13 mediated asthma phenotypes. TGF-β1 is believed to participate in myofibroblast phenotypic changes resulting in airway remodeling due to overexpression in IL-13 TG(+) mice (Lee et al., 2001). These findings suggest that TGF-β1 signaling pathways are negatively associated with AHR. We found that positivity in methacholine AHR was small, but statistically significant in subjects with the A allele in 2753G>A, whereas it was not significant in other alleles and in any haplotype. Therefore, genetic variations in this site may be partly associated with development of AHR.
The idea that Th2 cytokines can enhance AHR by promoting inflammatory cell recruitment remains popular (Venkayya et al., 2002). However, induction of AHR can occur in the absence of allergic airway inflammation (Ichinose et al., 2000). Our results suggest that variants of the TGFBR3 gene are associated with more positive effects on AHR than is the atopic status. Nevertheless, a study of TFG-β showed that plasma TGF-β1 levels were more elevated in stable non-atopic asthma than in atopic asthma (Joseph et al., 2003). This study showed that the statistical power of the association with nonatopic asthma was markedly greater for the Ht3 (TA) haplotype than for the 2753A allele alone. Although Th2 sensitization to allergens and airway inflammation are important in asthma, expression of asthma is also determined by the biologic functions of airway smooth muscle cells or myofibroblasts by inducing subepithelial and lung parenchymal fibrosis (Martin et al., 2000). In terms of the genetic effects of individual SNPs in the TGFBR3 gene on expression of AHR, we suggest that the A allele in 2753G>A is associated with asthma pathogenesis via airway hyperreactivity.
One important issue for genetic association studies is that an imprecise definition of a disease phenotype can cause discrepancies in results. For asthma association studies, variations of asthma phenotype definitions in genome-wide association studies are large and vary from loose definitions based only on clinical histories, to strict definitions based on clinical symptoms and objective parameters, such as reversible airway obstruction and AHR. In this study, asthma was strictly defined based on wheezing during the previous 12 months (as determined by questionnaire) and by the presence of a positive AHR. In terms of the genetic effects of SNPs of the TGFBR3 gene on asthma prevalence, this study showed that the prevalence of asthma, especially non-atopic asthma, was significantly greater in subjects with the Ht3 (TA) haplotype. However, this correlation was weakly associated with individual SNPs in the TGFBR3 gene, which suggests that haplotype analysis is more informative than individual SNP analysis. The association between the prevalence of asthma and genetic variations in the TGFBR3 gene was remarkable in non-atopic subjects, but not in atopic subjects. This finding suggests that genetic variations in the TGFBR3 gene are partly associated with development of non-atopic asthma.
In summary, the minor allele in 2753G>A and the haplotype 44T_2753A of the TGFBR3 gene were associated with the prevalence of asthma, especially non-atopic asthma. These findings suggest that genetic variations in TGF-β receptors are important genetic markers for predicting the development of asthma.
Methods
Subjects
A cohort of 2,118 ethnic Korean subjects (1,033 male and 1,085 female) aged from 10 to 18 years living in rural areas of Jeju Island, Korea was randomly recruited. Parents gave written informed consent, and the study protocol was approved by the Ethics Committee of Seoul National University Hospital. All subjects responded to a questionnaire concerning asthma symptoms and risk factors. The questionnaire, developed by the International Study of Asthma and Allergic disease in Children (ISAAC), was translated into Korean following guidelines developed by ISAAC (Kim et al., 2001). Questions concerning asthma symptoms were concentrated on a recurrent wheezing and a nocturnal cough in the absence of respiratory tract infection over the preceding 12 month period. Questions on risk factors covered a family history of allergic disease, a history of passive smoking, and the vaccination history, including measles, M. tuberculosis, and the hepatitis B virus.
None of the subjects had been treated or had used oral or inhaled bronchodilators for five days preceding methacholine bronchial provocation testing (MBPT). Subjects who had contracted an upper respiratory tract infection during the two week period prior to the study were excluded from the methacholine challenge. A total of 2,055 subjects underwent methacholine challenge, as previously described (Kim et al., 2001). Methacholine AHR was expressed as PC20, and regarded as positive if the PC20 was < 16 mg/ml.
To evaluate Th2 sensitization to allergens, serum total IgE levels were determined and skin prick testing against common aeroallergens was performed. Total serum IgE levels were determined in 2,058 subjects using ELISA (Kim et al., 1999). Subjects who had received oral antihistamines during the five days prior to skin prick testing or had dermographism were excluded from the skin prick testing. A total of 2,047 subjects underwent skin prick testing against 11 common aeroallergens (Allergopharma, Germany) as previously described (Kim et al., 2002). Atopy was defined as a positive skin prick test response (allergen/histamine ratio >1.0 plus a mean wheal size >4 mm) to one or more allergens. The skin index was defined as the number of positive skin tests. Asthma was defined when a subject had a current wheeze by questionnaire and showed a positive AHR. Atopic or non-atopic asthma was classified based on the presence of atopy.
Genotyping
Sequencing and SNP
After isolating genomic DNA from the peripheral blood of 24 healthy Korean subjects using a QIAamp DNA blood kit following the manufacturer's instruction (Qiagen, Hilden, Germany), 2 kb of the 5'-upstream region in the promoter and all exons including the exon - intron boundaries of TGFBR3 were amplified using PCR [Reference genome sequences; NM_000358 (TGFBI), NM_003243 (TGFBR3) released on 2 Mar 2006]. Amplified PCR products were sequenced using a Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City) in both directions according to standard protocols. After sequencing of TGFBR3, 19 genetic polymorphisms with minor allele frequencies of greater than 2% were identified. Of these, 2 SNPs [44T>C (S15F); a nonsynonymous SNP, and 2753G>A; at 3'UTR in TGFBR3] were selected for scoring after considering location (SNPs in exons and promoter region had priority), linkage disequilibrium (LD) patterns (only one SNP, if there were tight LDs), and the haplotype tagging status (Table 2).
SNP scoring
Selected SNPs were scored using the high throughput single base - pair extension method (SNP - IT™ assay) with a SNPstream25K system, which was customized to automatically genotype DNA samples in 384 well plates and to provide a colorimetric readout (Orchid Biosciences, New Jersey) (Han et al., 2004).
Statistics
Individual SNPs and haplotypes were analyzed as three component variables (for example, AA, AB, and BB; where A is the major frequency allele or haplotype and B is the minor frequency allele or haplotype). Haplotypes and their frequencies were estimated using an expectation maximization algorithm. To determine whether one of the homozygous genotypes had an effect that differed from the common effects of the heterozygous genotype or the other homozygote (i.e., AA vs. AB+BB or BB vs. AB+AA, dominant or recessive models, respectively), a 2 × 2 contingency table was constructed and P values were obtained using a χ2 test for categorical variables. To determine odds ratios (ORs) and 95% confidence intervals (CIs), multiple logistic regression modeling was performed to adjust for confounding variables, such as, age, sex, a family history of allergic disease, a history of passive smoking, and the vaccination history. However, in the case of serum total IgE levels, the log-transformed total IgE levels exhibited a right skew and, thus, statistical significance was evaluated using non-parametric methods. The Hardy - Weinberg equilibrium was analyzed using the χ2 test. All statistical analyses were performed using SAS software (version 8.1, Cary, NC). P values of < 0.05 were regarded as significant.
Abbreviations
- AHR:
-
airway hyperresponsiveness
- LD:
-
the linkage disequilibrium
- SNP:
-
single nucleotide polymorphism
- TGFBR:
-
transforming growth factor-β receptor
References
Blobe GC, Liu X, Fang SJ, How T, Lodish HF . A novel mechanism for regulating transforming growth factor beta (TGF-beta) signaling . J Biol Chem 2001 ; 276 : 39608 - 39617
Carsten B, Schmidt W, Kurt B . Regulation and role of transforming growth factor-β in immune tolerance induction and inflammation . Curr Opin in Immunol 2004 ; 16 : 709 - 716
Catherine D, Véronique F, Nelly F . Transforming growth factor-β and its role in asthma . Pulm Pharmacol Ther 2003 ; 16 : 181 - 196
Chen W, Wahl SM . TGF-beta: receptors, signaling pathways and autoimmunity . Curr Dir Autoimmun 2002 ; 5 : 62 - 91
Dijke P, Hill CS . New insights into TGF-β-Smad signalling . Trends Biochem Sci 2004 ; 29 : 265 - 273
Han W, Kang D, Park IA, Kim SW, Bae JY, Chung KW, Noh DY . Associations between breast cancer susceptibility gene polymorphisms and clinicopathological features . Clin Cancer Res 2004 ; 10 : 124 - 130
Holgate ST . Airway inflammation and remodeling in asthma: current concepts . Mol Biotechnol 2002 ; 22 : 179 - 189
Ichinose M, Takahashi T, Sugiura H, Endoh N, Miura M, Mashito Y, Shirato K . Baseline airway hyperresponsiveness and its reversible component: role of airway inflammation and airway calibre . Eur Respir J 2000 ; 15 : 248 - 253
Joseph J, Benedict S, Badrinath P, Wassef S, Joseph M, Abdulkhalik S, Nicholls MG . Elevation of plasma transforming growth factor beta1 levels in stable nonatopic asthma . Ann Allergy Asthma Immunol 2003 ; 91 : 472 - 476
Kim SY, Kim TB, Moon KA, Kim TJ, Shin DW, Cho YS, Moon HB, Lee KY . Regulation of pro-inflammatory responses by lipoxygenases via intracellular reactive oxygen species in vitro and in vivo . Exp Mol Med 2008 ; 40 : 461 - 476
Kim YK, Cho SH, Koh YY, Son JW, Jee YK, Lee MH, Min KU, Kim YY . Skin reactivity to inhalant allergens, total serum IgE levels, and bronchial responsiveness to methacholine are increased in parents of nonatopic asthmatic children . J Allergy Clin Immunol 1999 ; 104 : 311 - 316
Kim YK, Park HS, Kim HY, Jee YK, Son JW, Bae JM, Lee MH, Cho SH, Min KU, Kim YY . Citrus red mite (Panonychus citri) may be an important allergen in the development of asthma among exposed children . Clin Exp Allergy 2001 ; 31 : 582 - 589
Kim YK, Chang YS, Lee MH, Hong SC, Bae JM, Jee YK, Chun BR, Cho SH, Min KU, Kim YY . Role of environmental exposure to spider mites in the sensitization and the clinical manifestation of asthma and rhinitis in children and adolescents living in rural and urban areas . Clin Exp Allergy 2002 ; 32 : 1305 - 1309
Lee CG, Homer RJ, Zhu Z, Lanone S, Wang X, Koteliansky V . Interleukin - 13 induces tissue fibrosis by selectively stimulating and activating TGF-β1 . J Exp Med 2001 ; 194 : 809 - 821
Long AD, Langley DH . The power of association studies to detect the contribution of candidate genetic loci to variation in complex traits . Genome Res 1999 ; 9 : 720 - 731
Mak JC, Leung HC, Ho SP, Law BK, Ho AS, Lam WK, Ip MS, Chan-Yeung MM . Analysis of TGF-beta(1) gene polymorphisms in Hong Kong Chinese patients with asthma . J Allergy Clin Immunol 2006 ; 117 : 92 - 96
Martin JG, Duguet A, Eidelman DH . The contribution of airway smooth muscle to airway narrowing and airway hyperresponsiveness in disease . Eur Respir J 2000 ; 16 : 349 - 354
Mauviel A . Transforming growth factor-beta: key mediator of fibrosis . Methods Mol Med 2005 ; 117 : 69 - 80
Morén A, Ichijo H, Miyazono K . Molecular cloning and characterization of the human and porcine transforming growth factor - beta type III receptors . Biochem Biophys Res Commun 1992 ; 189 : 356 - 362
Mosmann TR, Cherwinski HM, Bond WM, Giedlin A, Coffman RL . Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins . J Immunol 1986 ; 136 : 2348 - 2357
Moustakas A, Pardali K, Gaal A, Heldin CH . Mechanisms of TGF-beta signaling in regulation of cell growth and differentiation . Immunol Lett 2002 ; 82 : 85 - 91
Nagpal K, Sharma S, B - Rao C, Nahid S, Niphadkar PV, Sharma SK, Ghosh B . TGF-beta1 haplotypes and asthma in Indian populations . J Allergy Clin Immunol 2005 ; 115 : 527 - 533
Risch N, Merikangas K . The future of genetic studies of complex human diseases . Science 1996 ; 273 : 1516 - 1517
Sankar S, Mahooti - Brooks N, Centrella M, McCarthy TL, Madri JA . Expression of transforming growth factor type III receptor in vascular endothelial cells increases their responsiveness to TGF-β2 . J Biol Chem 1995 ; 270 : 13567 - 13572
Shull MM, Ormsby I, Kier AB, Pawlowski S, Diebold RJ, Yin MAllen R, Sidman C, Proetzel B, Calvin D, Annunziata N, Doeschman T . Targeted disruption of the mouse transforming growth factor-beta1 gene results in multifocal imflammtory disease . Nature 1992 ; 359 : 693 - 699
Townley RG, Horiba M . Airway hyperresponsiveness-a story of mice and men and cytokines . Clin Rev Allergy Immunol 2003 ; 24 : 85 - 110
Venkayya R, Lam M, Willkom M, Grunig G, Corry DB, Erle DJ . The Th2 lymphocyte products IL-4 and IL-13 rapidly induce airway hyperresponsiveness through direct effects on resident airway cells . Am J Respir Cell Mol Biol 2002 ; 26 : 202 - 208
Wiebke S, Stefan B, Gesine H . Reduced expression of transforming growth factor β1 exacerbates pathology in an experimental asthma model . Eur J Immunol 2005 ; 35 : 198 - 206
Wiesch DG, Meyers DA, Bleecker ER . Genetics of asthma . J Allergy Clin Immunol 1999 ; 104 : 895 - 901
Wills-Karp M, Luyimbazi J, Xu X, Schofield B, Neben TY, Karp CL, Donaldson DD . Interleukin-13, central mediator of allergic asthma . Science 1998 ; 282 : 2258 - 2261
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Kim, HK., Jang, TW., Jung, MH. et al. Association between genetic variations of the transforming growth factor β receptor type III and asthma in a Korean population. Exp Mol Med 42, 420–427 (2010). https://doi.org/10.3858/emm.2010.42.6.043
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3858/emm.2010.42.6.043