Abstract
Mutations in the PCYT1A gene have been recently linked to two different phenotypes: one characterized by spondylometaphyseal dysplasia and cone-rod dystrophy (SMD-CRD) and the other by congenital lipodystrophy, severe fatty liver disease, and reduced HDL cholesterol without any retinal or skeletal involvement. Here, we identified, by next generation sequencing, sequence variants affecting function in the PCYT1A gene in three young patients with isolated retinal dystrophy from two different Italian families. A thorough clinical evaluation of the patients, with whole skeleton X-ray, metabolic assessment and liver ultrasound failed to reveal signs of skeletal dysplasia, metabolic and hepatic alterations. This is the first report showing that the PCYT1A gene can be responsible for isolated forms of retinal dystrophy, particularly without any skeletal involvement, thus further expanding the phenotypic spectrum induced by mutations in this gene.
Similar content being viewed by others
Introduction
Inherited retinal dystrophies (IRDs) are a heterogeneous group of conditions that represent the most frequent causes of visual impairment of genetic origin in the western world.1, 2 IRDs display a notable extent of genetic heterogeneity with more than 200 causative genes identified over the last few years (http://www.sph.uth.tmc.edu/RetNet/). Clinically, they can be either isolated or syndromic, that is, when there is extraretinal involvement. Isolated forms of IRDs include Retinitis pigmentosa (RP), Leber Congenital Amaurosis (LCA), Achromatopsia (ACHM), Cone (CD) and cone-rod dystrophies (CRD). There are several syndromic forms including the recently described spondylometaphyseal dysplasia with CRD (SMD-CRD).3
SMD-CRD is a rare autosomal recessive disorder with postnatal growth deficiency resulting in severe short stature, rhizomelia with bowing of the lower limbs, platyspondyly with anterior vertebral protrusions, progressive metaphyseal irregularity and cupping with shortened tubular bones. Affected patients have early-onset, progressive visual impairment associated with a pigmentary maculopathy and electroretinographic evidence of cone-rod dysfunction.3, 4, 5 SMD-CRD is caused by mutations in the PCYT1A gene,4, 5 which encodes the enzyme CTP: phosphocoline cytidyltransferase alpha (CCTα), the rate-determining enzyme in phosphatidylcholine (PC) biosynthesis (Kennedy pathway). PC is the main glycerophospholipid in eukaryotic cells and is an essential component of all cellular membranes.6 Besides SMD-CRD, PCYT1A mutations have also been linked to a distinct phenotype presenting with congenital lipodystrophy and fatty liver disease without any retinal involvement or signs of spondylometaphyseal dysplasia apart from a modest short stature.6
Subjects and methods
Patients
The reported patients were diagnosed at the Eye Clinic of the Multidisciplinary Department of Medical, Surgical and Dental Sciences, Università degli Studi della Campania ‘Luigi Vanvitelli’, and at the Section of Pediatrics, Department of Translational Medicine, Federico II University, Naples, Italy.
Ethical approval for this study was obtained by the Università degli Studi della Campania ‘Luigi Vanvitelli’ Medical Ethics Committee.
Methods
Genomic DNA, extracted from the peripheral blood of the patients, was used for both targeted and Whole Exome Sequencing by Next Generation Sequencing (NGS) procedures. For the targeted analysis, target enrichment was obtained with the HaloPlex Target Enrichment System (Agilent Technologies, Santa Clara, CA, USA) using previously described procedures.7, 8 For WES library preparation, we used the Sure Select QXT Clinical Research Exome kit following the manufacturer’s instructions (Protocol Version D0, November 2015, Agilent Technologies). Sequencing libraries were then sequenced using the Next Seq500 system performing paired-end runs covering at least 2 × 150 nucleotides (nt) (Illumina Inc., San Diego, CA, USA). Search for sequence variants putatively affecting function was carried out, both for the targeted and for the WES analysis, using previously described approaches.7 More specifically, we selected non-synonymous single nucleotide variations (SNVs) and indels, with a frequency lower than 1% in public databases as well as in our internal database containing WES data from 300 Italian subjects. For paternity test in individual A322, we used the AmpFlSTR Profiler Plus Kit (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA).
PCYT1A sequence variants described in this report have been uploaded to Leiden Open Variation Database (LOVD v3.0, http://databases.lovd.nl/shared/variants) with the following variant IDs: 0000149994, 0000149995, 0000149996, 0000149997. Reference sequences used were: RefSeq NM_005017.2 (cDNA), NG_042817.1 (genomic), NP_001299602.1 (protein) and EC 2.7.7.15 (enzyme).
Results and discussion
In the course of a project aimed at the identification of the genetic basis of IRDs by Next Generation Sequencing (NGS) in 40 Italian families (Banfi and Simonelli, manuscript in preparation), we analyzed three patients, from two independent families. The patients were a male, A322, from one family and two sisters, A333a and A333b, from another family. All of them had previously received a diagnosis of LCA.9
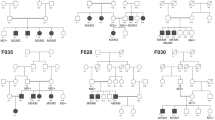
We first performed a targeted NGS analysis on the genomic DNA of patients A322 and A333a using a panel, named RETplex, covering the coding exons of 140 known IRD genes (Banfi and Simonelli, manuscript in preparation). We obtained a high coverage in both patients with an average coverage depth of 100 × for over 90% of the RETplex target. The only putatively pathogenic variants, under a presumed recessive inheritance and using the filtering criteria reported in Methods, were identified in the PCYT1A gene in both patients (Figure 1a). In particular, we detected a heterozygous splice site variant (c.897+1G>A) and a heterozygous missense variant (c.277G>A, p.(A93T)) in patient A322. Patient A333a was heterozygous for the c.277G>A (p.(A93T)) missense variant and for the nonsense variant c.847C>T (p.(R283*)). All the sequence variants were validated by Sanger sequencing. Segregation analysis of the variants in both families showed that they were in trans in all three patients (Figure 1a). In Family 1, the patient’s mother and his healthy brother were found to be heterozygous carriers of the c.277G>A (p.(A93T)) variant whereas the father displayed a wild-type genotype for both alleles (Figure 1a). This suggests that the c.897+1G>A variant in patient A322 arose as a de novo event, as non-paternity was ruled out using the AmpFlSTR® Profiler Plus Kit (Supplementary Table S1). To determine the parental origin of this de novo event, we subcloned a genomic DNA fragment, obtained by PCR, from patient A322, spanning both the c.897+1G>A variant and the rs3184886 SNP, which was present in the heterozygous state both in the patient and in his father while it was absent in the mother (data not shown). Sanger sequencing of several subclones (n=10) corresponding to the separated parental alleles revealed that the rs3184886 SNP was in cis with respect to the c.897+1G>A variant thus confirming its paternal origin. Please note that we did not find in patient A322 any heterozygous variant putatively affecting function in the CRX and IMPDH1 genes, which were previously linked to dominant forms of LCA. In family 2, patient A333b was also found to bear the same PCYT1A variants present in her sister A333a whereas each of the two parents was heterozygous carrier of only one of them. None of the above variants were found in the Exome Variant Server (http://evs.gs.washington.edu/EVS/), in the ExAC Browser (http://exac.broadinstitute.org) and in our internal database including over 300 whole exomes from Italian subjects. The variant c.847C>T (p.(R283*)) was previously reported as likely affecting function in patients with SMD-CRD.4 The splice variation c.897+1G>A is likely to be a loss-of-function variant and the c.277G>A, p.(A93T)) missense variant affects an alanine residue extremely conserved across evolution, which resides in the catalytic domain (Figure 1b) and is predicted to be deleterious by Polyphen210 and by MutationTaster.11
New PCYT1A variants associated with isolated retinal dystrophy and their distribution within the protein. (a) Pedigrees of the two families described in this report along with the segregation of their PCYT1A mutant genotypes. =, wild-type allele. (b) Multiple sequence alignment of part of the PCYT1A catalytic domain across metazoa. The red arrow marks the highly conserved alanine residue that is affected by the c.277G>A, p.(A93T)) missense variant identified in patients A322, A333a and A333b. Hsa, Homo sapiens, NP_005008; Mmu, Mus musculus, NP_001156631; Gga, Gallus gallus, XP_422725, Xtr, Xenopus tropicalis, XP_002934091; Dre, Danio rerio, NP_001017634; Dme, Drosophila melanogaster, NP_647622; Cel, Caenorhabditis elegans, NP_001033539. (c) Location of all PCYT1A mutations identified so far with respect to known functional domains. N, N-terminal domain (orange), also including the nuclear localization signal; C, catalytic domain (light blue); M, membrane binding domain (yellow); P, C-terminal phosphorylated domain (green). The different PCYT1A phenotypes corresponding to the mutations shown in the diagram are highlighted with different colors. No apparent genotype-phenotype correlation can be identified. SMD-CRD, spondylometaphyseal dysplasia with cone-rod dystrophy (dark blue): mutations reported in;4, 5 CLD-FL, congenital lipodystrophy and severe fatty liver disease (light green): mutations reported in;6 LCA, (light brown): variants described in this report.
To further corroborate the likely pathogenic role of the above-described PCYT1A variants, we also carried out Whole Exome Sequencing (WES) analysis in subjects A333a and A322. For variants filtering, we used the same criteria utilized for the targeted sequencing (see Methods). WES analysis led to the identification of 3 sequence variants likely affecting functions in 2 different genes in subject A333a and 6 in 4 different genes in subject A322 under an autosomal recessive pattern of inheritance (Supplementary Table S2). WES analysis further strengthened the likely pathogenic role of PCYT1A variants both because this genes was, among all the other candidates, the most highly correlated to the phenotype observed in the patients and also due to its significant expression levels in the human retina.12 On the basis of all of the above observations we concluded that sequence variants in the PCYT1A gene are responsible for the clinical conditions present in patients A322, A333a and A333b.
Since mutations in PCYT1A have been associated with two different clinical conditions, namely SMD-CRD4, 5 and to congenital lipodystrophy with fatty liver disease6 (Figure 1c), we performed a detailed clinical evaluation of all three patients to rule out the presence of the abnormalities that were previously reported in patients harboring PCYT1A mutations.
Family 1
Patient A322 is the first of two sons of healthy non-consanguineous parents and his brother is healthy and has no retinal abnormalities, as assessed by ophthalmological evaluation. At the age of conception of the proband the mother was 34 and the father was 40 years old. The patient was born at term of gestation with normal length, weight and head circumference. There were no clinical problems until he was 2 years old and he was noted to have night-blindness and reduced visual acuity. At the age of 4 years, he showed a best-corrected visual acuity (BCVA), measured using the LEA symbols, of 20/200 in both eyes (right eye (RE), sphere +1,50 =cylinder +1 alpha (axis) 90°, left eye (LE), sphere +1,50=cylinder +2 alpha 75°), normal color vision and ocular motility. Lenses were clear. Fundus examination revealed the presence of pink optic disc, widespread retinal pigment epithelium dystrophy with areas of hypo- and hyperpigmentation, and normal retinal vessels (Figure 2a). Optical coherence tomography (OCT) examination, performed with the spectral domain OCT (SD-OCT) (Cirrus HD-OCT; Carl Zeiss, Dublin, California, USA), showed reduced macular thickness (Mean macular thickness (MMT) RE 110 μm; LE 100 μm; normal value 212±20 μm) with mild retinal pigment epithelium (RPE) dystrophy (Figure 2b). Fundus autofluorescence (AF) showed a widespread, mild hypoautofluorescence (Supplementary Figure 1A). Electroretinographic (ERG) analysis revealed the presence of scotopic and photopic traces below noise level.
Ophthalmological findings in patients A322, A333a and A333b. A322 (a) retinography: presence of pink optic disk, widespread retinal pigment epithelium dystrophy with areas of hypo- and hyperpigmentation, normal retinal vessels; (b) OCT: reduced macular thickness with mild retinal pigment epithelium (RPE) dystrophy. A333b (c) retinography: presence of pink optic disc, pigment deposits resembling bone spicules in mid periphery, normal retinal vessels; (d) OCT: reduced macular thickness with RPE dystrophy. A333a (e) retinography: presence of pigment deposits resembling bone spicules in mid periphery; (f) OCT: reduced macular thickness with severe RPE dystrophy. (g) retinography of an healthy eye. (h) OCT of an healthy eye: all retinal layers are labeled.
At 4 years, that is, the age of last observation, the patient had normal height (107.5 cm, 75th–90th centile), weight (18.50 kg, 75th–90th centile), and head circumference (50.5 cm, 50th centile). No facial dysmorphisms were noted, and body proportions and range of motion of the joints were normal. There was no evidence of lipodystrophy on skin exam. His cognitive development and hearing were normal. Routine hematological and biochemical tests yielded normal results (including, among others, AST 20 UI/l, ALT 13 UI/l, GGT 13UI/l, total bilirubin 0.7 mg/dL, fasting glycemia 89 mg/dL and fasting insulinemia 15 mUI/ml). Lipid profile was also normal (total cholesterol 165 mg/dL, HDL 55 mg/dL, tryglicerides 88 mg/dL).
Whole skeletal X-ray revealed no signs of spondylometaphyseal dysplasia but coccyx agenesis was noted. A delayed bone age (2 years and 8 months) was also noted. Liver ultrasound showed no signs of fatty infiltration and normal size.
Family 2
Patients A333a and A333b are two daughters of healthy, non-consanguineous parents. Both patients were born at term with normal height, weight, and head circumference. Growth and neurocognitive development were normal.
Patient A333a underwent her first ophthalmological evaluation at 8 months of age because of nystagmus and her visual function progressively decreased over time. At the age of 19 years, she showed a BCVA of 20/1000 in both eyes and exotropia. Lenses were clear. Fundus examination revealed a pink optic disc, RPE dystrophy with rare pigment deposits resembling bone spicules in mid periphery and normal retinal vessels (Figure 2e). OCT examination showed reduced macular thickness (MMT in RE 176 μm; LE 120 μm) with severe RPE dystrophy (Figure 2f). AF showed a widespread hypoautofluorescence with no autofluorescence macular area (Supplementary Figure 1b). Goldmann visual field showed patchy losses of peripheral vision. ERG revealed the presence of scotopic and photopic traces below noise level.
At her latest evaluation, at the age of 19 years, the patient had height (152 cm, 5th centile), weight (63.5 Kg, 50th–75th centile), and head circumference (53 cm; between average and −2 SD) within normal ranges. She was non-dysmorphic and had no signs of lipodystrophy. Her cognitive functions and hearing were normal. Routine hematological and biochemical tests were all normal (AST 19 UI/l, ALT 18 UI/l, GGT 23 UI/L, total bilirubin 0.5 mg/dl, fasting glycemia 74 mg/dl, fasting insulinemia 9.5 mUI/ml). Lipid profiles were also normal (total cholesterol 183 mg/dl, HDL 54 mg/dl, tryglicerides 69 mg/dl).
The skeletal X-ray showed no signs of spondylometaphyseal dysplasia. On ultrasound, the liver was normal although it was slightly increased in size (longitudinal dimension of right lobe 151.6 mm, left lobe 97 mm, caudate lobe 47 mm).13
Patient A333b was first referred to ophthalmological examination at the age of 1 year because of nystagmus. At the age of 12 years, she showed a BCVA of 20/400 in both eyes. Fundus examination revealed a pink optic disc, RPE dystrophy with rare pigment deposits resembling bone spicules in mid periphery and normal retinal vessels (Figure 2c). OCT examination showed reduced macular thickness (MMT 103 μm in both eyes) with RPE dystrophy (Figure 2d). AF showed a widespread hypoautofluorescence and no autofluorescence in macular area (Supplementary Figure 1C). Goldmann visual field showed central 10° preserved. ERG revealed the presence of scotopic and photopic traces below noise level.
At her last observation at the age of 12 years, the patient had normal height (145 cm, 10th-25th centile), weight (46.5 Kg, 50th- 75th centile), and head circumference (54 cm; between average and +2 SD). She had no facial dysmorphisms and no abnormalities of the skin. Her cognitive functions and hearing were normal. Routine hematological and biochemical tests yielded normal results, including, among others, AST 20 UI/l, ALT 13 UI/l, GGT 13UI/l, total bilirubin 0.7 mg/dl, fasting glycemia 89 mg/dl, fasting insulinemia 15 mUI/ml. Lipid profiles were normal (total cholesterol 165 mg/dl, HDL 55 mg/dl and tryglicerides 88 mg/dl).
The skeletal survey revealed no significant bone abnormalities but only a mild dorsal–lumbar scoliosis. The liver ultrasound showed no abnormalities and the liver had normal size.
Altogether, based on the above observation, patients A322, A333a and A333b are affected by an isolated form of IRD without evidence of extraretinal involvement. They display an overall similar retinal phenotype that can be classified as LCA.9 All three patients had an early-onset severe visual impairment; two of them presented nystagmus within the first year of age. The fundus appearance ranged from ‘salt and pepper’ dystrophy to pigmentary deposits resembling bone spicules. In all three patients, macular thickness was reduced and ERG amplitude was below noise level. We therefore conclude that mutations in the PCYT1A gene can be associated not only with Cone-rod dystrophy (CRD) as previously described4, 5 but also with LCA. There are already examples of genes whose mutations can either be linked to CRDs or to LCA, such as in the case of CRX and GUCY2D.14 Our findings not only highlight the value of using comprehensive targeted gene panels for a precise molecular characterization of IRD patients but further support the necessity to integrate clinical and genetic data in order to ensure the proper management of this heterogeneous group of conditions.
Notably, none of the three patients described here showed any sign of spondylometaphyseal dysplasia previously described in patients with mutations in PCYT1A. The only skeletal abnormalities found included coccyx agenesis, delayed bone age, and mild scoliosis that are relatively non-specific findings and appear unlikely related to the PCYT1A genotype.
Mutations in the PCYT1A gene have also been associated with congenital lipodystrophy and severe fatty liver and low HDL cholesterol levels in two patients without ophthalmological or skeletal involvement although mild short stature was reported in one affected individual.6 Although a mild increase in liver size was detected in one of our patients, none of the three patients showed signs of hepatic steatosis, lipodystrophy, insulin resistance or abnormalities of lipid profiles. However, all our patients were children or young adults and we cannot rule out that hepatic or metabolic alterations may become evident at later ages. For this reason, we are planning to monitor our patients for these abnormalities over time. Nevertheless, this represents the first evidence of PCYT1A mutations that are responsible for a retinal phenotype that is not accompanied by spondylometaphyseal dysplasia or other significant skeletal abnormalities.
The sequence variants identified in patients A322, A333a and 333b do not seem to cluster differently from those reported to be associated with the previously described PCYT1A-related phenotypes (Figure 1c). Interestingly, the previously unreported c.277G>A, p.(A93T)) variant was found in all three of the patients from two independent families with isolated IRD described here. However, this variant falls within the catalytic domain of the PCYT1A protein that harbors several other missense mutations already found in patients with different phenotypes (Figure 1c). A striking example is provided by three sibs from a North European family (reported as family 4) described by Hoover-Fong et al,4 with clear signs of SMD-CRD that were reported to share one sequence variant (namely, p.(R283*)) with family 2 in this report. Moreover, the two families display different missense mutations in the catalytic domain that are located only six residues apart (c.277G>A, p.(A93T)) in the case of Family 2 of this report and p.(A99T) in the case of Family 4 reported by Hoover-Fong et al.4). Therefore, as also previously suggested,4, 6 the phenotypic heterogeneity of PCYT1A mutations does not seem to be explained by variants affecting specific domains of the protein but rather genetic modifiers may play a role in the broad phenotypic spectrum caused by PCYT1A mutations.
In summary, this is the first report showing that sequence variants in the PCYT1A gene can be responsible for isolated forms of LCA, thus expanding the phenotypic spectrum induced by mutations in this gene. We therefore suggest that the pathogenic role of PCYT1A should be evaluated in patients with IRD even in the absence of skeletal anomalies.
References
Berson EL : Retinitis pigmentosa. The Friedenwald Lecture. Invest Ophtalmol Vis Sci 1993; 34: 1659–1676.
Dryja TP, Li T : Molecular genetics of retinitis pigmentosa. Hum Mol Genet 1995; 4: 1739–1743.
Walters BA, Raff ML, Hoeve JV et al: Spondylometaphyseal dysplasia with cone-rod dystrophy. Am J Med Genet A 2004; 129A: 265–276.
Hoover-Fong J, Sobreira N, Jurgens J et al: Mutations in PCYT1A, encoding a key regulator of phosphatidylcholine metabolism, cause spondylometaphyseal dysplasia with cone-rod dystrophy. Am J Hum Genet 2014; 94: 105–112.
Yamamoto GL, Baratela WA, Almeida TF et al: Mutations in PCYT1A cause spondylometaphyseal dysplasia with cone-rod dystrophy. Am J Hum Genet 2014; 94: 113–119.
Payne F, Lim K, Girousse A et al: Mutations disrupting the Kennedy phosphatidylcholine pathway in humans with congenital lipodystrophy and fatty liver disease. Proc Natl Acad Sci USA 2014; 111: 8901–8906.
Di Fruscio G, Schulz A, De Cegli R et al: Lysoplex: An efficient toolkit to detect DNA sequence variations in the autophagy-lysosomal pathway. Autophagy 2015; 11: 928–938.
Savarese M, Di Fruscio G, Mutarelli M et al: MotorPlex provides accurate variant detection across large muscle genes both in single myopathic patients and in pools of DNA samples. Acta Neuropathol Commun 2014; 2: 100.
Chacon-Camacho OF, Zenteno JC : Review and update on the molecular basis of Leber congenital amaurosis. World J Clin Cases 2015; 3: 112–124.
Adzhubei IA, Schmidt S, Peshkin L et al: A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249.
Schwarz JM, Cooper DN, Schuelke M, Seelow D : MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 2014; 11: 361–362.
Pinelli M, Carissimo A, Cutillo L et al: An atlas of gene expression and gene co-regulation in the human retina. Nucleic Acids Res 2016.
Konus OL, Ozdemir A, Akkaya A, Erbas G, Celik H, Isik S : Normal liver, spleen, and kidney dimensions in neonates, infants, and children: evaluation with sonography. AJR Am J Roentgenol 1998; 171: 1693–1698.
Walia S, Fishman GA, Jacobson SG et al: Visual acuity in patients with Leber's congenital amaurosis and early childhood-onset retinitis pigmentosa. Ophthalmology 2010; 117: 1190–1198.
Acknowledgements
We thank the participating patients and their families. We are grateful to Brunella Franco for critical reading of the manuscript and Carmela Acerra for text editing. We thank Francesca Del Vecchio Blanco for technical assistance. We also thank the TIGEM Next Generation Sequencing facility for support in targeted sequencing; Francesco Musacchia and the TIGEM Bioinformatics Core for support in NGS data analysis. This work was supported by grants from the Italian Fondazione Roma (to SB and FS) and from the Italian Telethon Foundation (to SB).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Testa, F., Filippelli, M., Brunetti-Pierri, R. et al. Mutations in the PCYT1A gene are responsible for isolated forms of retinal dystrophy. Eur J Hum Genet 25, 651–655 (2017). https://doi.org/10.1038/ejhg.2017.23
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2017.23