Abstract
The POT1 protein binds and protects telomeres. Germline variants in the POT1 gene have recently been shown to be associated with risk of developing tumors in different tissues such as familial chronic lymphocytic leukemia, colorectal, glioma and melanoma tumors. Recently, we uncovered a variant in the POT1 gene (p.R117C) as causative of familial cardiac angiosarcomas (CAS) in Li-Fraumeni-like (LFL) syndrome families. Our in silico studies predicted that this protein had lost the ability to interact with TPP1 and single-stranded DNA. In vitro studies corroborated this prediction and showed that this lack of function leads to abnormally long telomeres. To better understand the POT1 gene and its role with tumorigenesis, we extended the study to LFL (with and without members affected with angiosarcomas (AS)) and sporadic AS and cardiac sarcomas. We found POT1 variants in the 20% of the families with members affected with AS and 10% of sporadic AS and sarcomas. In silico studies predicted that these new variants were damaging in the same manner as previously described for the POT1 p.R117C variants. The wide spectrum of variants in the POT1 gene leading to tumorigenesis in different tissues demonstrates its general importance. Study of the POT1 gene should be considered as routine diagnostic in these cancers.
Similar content being viewed by others
Introduction
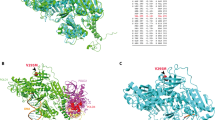
POT1 (protection of telomeres 1) is a component of the so-called shelterin complex, which binds and protects telomeres.1 POT1 binds TPP1; in turn, TPP1 binds the TRF1/2 proteins (Figure 1a). Two other conserved domains of the POT1 protein (oligonucleotide/oligosaccharide-binding 1 and 2; OB1 and OB2) directly interact with the telomere. The OB-fold is composed with the residues p.146–p.152. The stacking residues T1, T2, A3, G4, G5, G6, T7, T8, A9 and G10 interact with the single-stranded (ss) telomeric DNA sequence TTAGGGTTAG2 (Figure 1b).
Telomere biology. (a) Left: POT1 protein binds to TPP1 and to the ssDNA to regulate telomere maintenance. Right: individuals carrying the POT1 p.R117C variant showed reduced levels of POT1 bound to telomeres and to TPP1, which correlated with abnormally long, fragile telomeres and increased number of damage foci (γH2AX).9 (b) POT1 protein. OB1 and OB2 domains (N-ter) directly interact with the telomere through the stacking residues T1, T2, A3, G4, G5, G6, T7, T8, A9 and G10 (gray arrowheads). The OB-fold (position from p.146 to p.152) is shown (white rectangle). In the putative POT1 p.R117C protein, the PACC score of positions p.152 (OB-fold) and p.266 (T8; A9) changed from exposed (POT1) to buried (POT1 p.R117C), which lost the ability to bind to telomere. POT1 binds to TPP1 through the conserved domain located at the C terminus (TPP1). The POT1 p.R117C protein lost the ability to bind TPP1 due to the loss of the protein-binding site at position p.499 (white arrow). Variants in the POT1 gene found in this work (blue arrowheads) and in a previous sporadic CAS10 (green arrowhead) are shown. Variants in melanoma tumors4, 5 (white arrowheads), in glioma tumors3 (black arrowhead), in CLL7 (yellow arrowhead) and in colorectal cancer6 (red arrowhead) are also shown. TPP1, tripeptidyl peptidase-1 domain.
Germline variants in the POT1 gene have been described to be responsible for familial glioma,3 melanoma4, 5 and colorectal cancer.6 Germline and somatic variants in the POT1 gene were also described to be associated with chronic lymphocytic leukemia (CLL).7, 8 Recently, we identified a deleterious missense germline variant (rs780936436) in the POT1 gene (p.R117C), which caused cancer in three Li-Fraumeni-like (LFL) families, including members affected with cardiac angiosarcomas (CAS) and breast angiosarcoma (AS)9 (Figure 1b). A constitutional variant in the POT1 gene (p.R432*) was also found in one out of five sporadic CAS tumors10 (Figure 1b). CAS tumor is diagnosed in advanced stages when distant metastases are present and the survival is very poor, as surgical resection is not effective.
In silico studies suggested that the POT1 p.R117C protein had lost the ability to interact with ssDNA and TPP1 (Figure 1a).9 In vitro studies confirmed these in silico predictions and indicated that carriers of this variant had reduced levels of POT1 bound to the telomere and to the TPP1 protein, which correlated with abnormally long telomeres with increased fragility9 (Figure 1a).
To better understand the role of POT1 as one of the main genes responsible for the development of different familial cancer types, we extended our study to 34 TP53-negative LFL families (10 with and 24 without individuals affected with AS, respectively) and 30 cases of sporadic AS and cardiac sarcomas.
Materials and methods
DNA from peripheral blood and formalin-fixed paraffin-embedded tissues from a total of 64 patients with different tumors were selected for the whole POT1 gene study (see Supplementary Material for methods and patients details). Research Ethics Committee from Hospital Universitario de Fuenlabrada approved this study and written informed consent was obtained from all participants.
Reference sequences NM_015450.2 and NP_056265.2 are used for POT1 gene and POT1 protein, respectively. The number of the exons and introns from the POT1 gene is according the refseq: NG_029232.1. The variants described in this work have been submitted in the LOVD database freely available at www.LOVD.nl/POT1 (patient IDs: 105406–105411). In silico prediction software is detailed in Supplementary Material.
Results
Sequencing of the entire CDS of the POT1 gene (refseq: NM_015450.2) in the different series uncovered four new variants (one nonsense and three missense variants) and the same variant described in Calvete et al.9 (Table 1). The missense variants were all located within functional domains (refseq: NP_056265.2) and they were considered damaging by the functional predictors (Supplementary Materials) and the tolerance to amino-acid change score (Table 1).
Regarding the LFL families with members affected with AS, two variants were found in the POT1 gene. However, no variants in the POT1 gene were found in any individual of the 24 studied LFL families without individuals affected with AS. The missense p.(T497L) variant (c.1490C>A) was found in an individual affected with breast AS from a LFL French family (Table 1; Supplementary Figure S1). Putative protein–protein-binding sites were calculated for the putative protein containing the variant. The protein–protein-binding site at position p.499 was lost in this putative protein, as previously observed for the POT1 p.R117C protein. This resulted in the loss of capability of the mutant protein to interact with the TPP1 protein (Supplementary Table S1; Figure 1b). This prediction was confirmed by in vitro assays.9 The nonsense p.(Gln301*) variant (c.1560C>T) was found in an individual affected with CAS from another family (Table 1; Supplementary Figure S1). Binding capability to TPP1 of the putative POT1 protein with the p.(Gln301*) variant was not evaluated in silico because the entire TPP1 binding domain was truncated (Figure 1b).
In the series of individuals affected with sporadic tumors, the missense p.(P116L) variant (c.946C>T) was found in an individual with CAS (Table 1). The same protein-binding site lost at position p.499 in the POT1 p.R117C protein was observed in the putative POT1 p.(P116L) protein (Supplementary Table S1). In addition, the putative POT1 p.(P116L) protein changed the orientation from exposed (wt) to buried (PACC score for solvent accessibility) for two residues in the OB-fold and interacting with the ssDNA (p.152 and p.266), as previously described for the POT1 p.R117C protein (Figure 1b; Supplementary Table S2). Therefore, the POT1 p.(P116L) protein is also predicted to have lost its capacity to bind ssDNA. The series of individuals affected with sporadic tumors also included two cardiac sarcoma patients, one carrying the previously described p.(R117C) variant (c.948C>T) and another one with the intronic c.547-1G>A (CDS) variant (c.1145-1G>A) (Table 1). The intronic variant was located in the splice acceptor site of the eighth intron (NG_029232.1) and might lead to skipping of the site for splicing. The splice acceptor score was calculated for the wild-type POT1 DNA (score: 0.61; acceptance threshold: 0.45; Supplementary Figure S2). POT1 c.547-1G>A putatively lost the acceptor site (score: 0.00; acceptance threshold: 0.45), which would result in the skipping of the splice acceptor site. Therefore, removal of eighth intron of POT1 c.547-1G>A putatively leads to removal of ninth exon as well (NG_029232.1), which corresponds to the fifth exon of the CDS (NP_056265.2) (Supplementary Figure S2). The in silico studies with the putative POT1 c.547-1G>A protein were performed assuming that the entire fifth exon was lost. In the putative protein containing the POT1 c.547-1G>A variant, the site at p.499 for binding TPP1 was also predicted to be lost (Figure 1b; Supplementary Table S1; no RNA was available).
In summary, two different variants in the POT1 gene were found in LFL families with individuals affected with AS. In addition, another three variants in the POT1 gene were found in three individuals affected with sporadic tumors, one in a sporadic CAS patient and two in individuals affected with sporadic cardiac sarcomas.
LOH study was performed for the three individuals affected with sporadic tumors, carrying variants in the POT1 gene and available PFFE (Supplementary Table S3). No LOH was observed in the three studied tumors. This result was according our previous observation, where LOH was neither observed for the POT1 p.R117C variant.9
Discussion
Variants in the POT1 gene
Four new predicted damaging variants (p.(R116C), p.(Gln301*), p.(T497L) and c.547-1G>A; Table 1) have been added to the variant spectrum of the POT1 gene.
In silico studies predicted that these new variants are expected to be unable to bind to TPP1 as described for the POT1 p.R117C variant9 (Supplementary Table S1). In addition, the variant p.(P116L) is predicted to be defective in its interaction with ssDNA (Supplementary Tables S2). In vitro studies of the POT1 p.R117C protein substantiated the in silico prediction;9 therefore, carriers of the new variants described in the current study are expected to deregulate POT1 function in the same manner and to increase telomere length, making them unstable. Variants in telomere structure and maintenance genes lead to telomere fragility, which is commonly associated with different cancer types.11 Replication of the in silico results previously described for the in vitro-evaluated p.R117C variant, correlates with the association of developing tumors of the variants described in this work.
POT1 and AS
In the present work, variants in the POT1 gene were found in 2 out of the 10 studied LFL families, including members affected with AS (Supplementary Figure S1), but no variants were found in 24 LFL families without individuals affected with AS. Variants in the POT1 gene were also found in 3 out of the 30 individuals affected with sporadic tumors (2 in cardiac sarcomas and 1 in CAS; Table 2a). These variants are also important because they demonstrate that POT1 variants not only cause AS but also sarcomas in cardiac tissue.
Because cardiac tumors are rare, we added previously published cases.9, 10 Therefore, Table 2b includes 12 additional LFL families with members affected with AS, four of them had the POT1 p.(R117C) variant. Ten additional LFL families with members affected without AS did not present POT1 variants.9 Regarding sporadic CAS patients, one out of five additional patients presented a nonsense variant (p.R432*) in the POT1 gene.10 On the basis of these data, we can conclude that variants in the POT1 gene are present in 27.3% and 11.4% of LFL families with members affected with AS and sporadic CAS tumors, respectively (Table 2b).
POT1 variants in other diseases
Other described POT1 variants associated with risk of developing familial glioma and familial melanoma tumors also led to abnormally long telomeres.3, 4, 5 No information regarding telomere length was available for the familial CLL or colorectal cancer patients carrying POT1 variants.6, 7
All these variants described in different types of tumors appear randomly distributed along the gene and the conserved domains, independently of the cancer type (Figure 1b). Table 2c summarizes the frequency of alterations in these diseases. Although the incidence of POT1 variants is lower than in AS cases, they should be considered especially in familial melanoma (2.4%) and familial and sporadic CLL (6.1% and 3.5%, respectively).
In summary, we observed that variants in the POT1 gene are not limited to familial AS, but also occur in sporadic AS and cardiac sarcomas. POT1 variants are described mainly in cardiac tissue pathologies and a putative relation between cardiac tumors and malfunction of telomere biology might exist. However, the molecular landscape that leads to tumorigenesis is still not well understood. The wide spectrum of variants in the POT1 gene leading to tumorigenesis in different tissues demonstrates its general importance. Inclusion in cancer panels should be performed as routine diagnostic to provide earlier diagnosis of people at risk.
References
Palm W, de Lange T : How shelterin protects mammalian telomeres. Annu Rev Genet 2008; 42: 301–334.
Lei M, Podell ER, Cech TR : Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol 2004; 11: 1223–1229.
Bainbridge MN, Armstrong GN, Gramatges MM et al: Germline mutations in shelterin complex genes are associated with familial glioma. J Natl Cancer Inst 2014; 107: dju384.
Robles-Espinoza CD, Harland M, Ramsay AJ et al: POT1 loss-of-function variants predispose to familial melanoma. Nat Genet 2014; 46: 478–481.
Shi J, Yang XR, Ballew B et al: Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat Genet 2014; 46: 482–486.
Chubb D, Broderick P, Dobbins SE et al: Rare disruptive mutations and their contribution to the heritable risk of colorectal cancer. Nat Commun 2016; 7: 11883.
Speedy HE, Kinnersley B, Chubb D et al: Germline mutations in shelterin complex genes are associated with familial chronic lymphocytic leukemia. Blood 2016; 128: 2319.
Ramsay AJ, Quesada V, Foronda M et al: POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat Genet 2013; 45: 526–530.
Calvete O, Martinez P, Garcia-Pavia P et al: A mutation in the POT1 gene is responsible for cardiac angiosarcoma in TP53-negative Li–Fraumeni-like families. Nat Commun 2015; 6: 8383.
Kunze K, Spieker T, Gamerdinger U et al: A recurrent activating PLCG1 mutation in cardiac angiosarcomas increases apoptosis resistance and invasiveness of endothelial cells. Cancer Res 2014; 74: 6173–6183.
Karami S, Han Y, Pande M et al: Telomere structure and maintenance gene variants and risk of five cancer types. Int J Cancer 2016; 139: 2655–2670.
Acknowledgements
GB’s lab is supported by the INCa, the French National Cancer Institute and the ARC Foundation for Cancer Research. CL’s lab is funded by the Asociación Española Contra el Cáncer, Spanish Health Research Foundation, Carlos III Health Institute (PIE13/00022 and PI16/00563), Catalan Health Institute and Autonomous Government of Catalonia. AB’s lab is funded by Deutsche Forschungsgemeinschaft (BR1238/8-2). JB’s lab is partially funded by CIBERER, H2020 BRIDGES project and by the Spanish Ministry of Health supported by FEDER funds (PI16/00440).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Calvete, O., Garcia-Pavia, P., Domínguez, F. et al. The wide spectrum of POT1 gene variants correlates with multiple cancer types. Eur J Hum Genet 25, 1278–1281 (2017). https://doi.org/10.1038/ejhg.2017.134
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2017.134
This article is cited by
-
Spatial transcriptomics reveal topological immune landscapes of Asian head and neck angiosarcoma
Communications Biology (2023)
-
The European Journal of Human Genetics is turning 30: a selection of major cancer genetics papers published by the Journal
European Journal of Human Genetics (2022)
-
A Novel CHEK2 Variant Identified by Next-Generation Sequencing in an Italian Family with Li-Fraumeni Syndrome: Case Report
SN Comprehensive Clinical Medicine (2022)
-
Gametes deficient for Pot1 telomere binding proteins alter levels of telomeric foci for multiple generations
Communications Biology (2021)
-
Genetic predisposition for multiple myeloma
Leukemia (2020)