Abstract
In a consanguineous Turkish family diagnosed with autosomal recessive nonsyndromic hearing impairment (arNSHI), a homozygous region of 47.4 Mb was shared by the two affected siblings on chromosome 6p21.1-q15. This region contains 247 genes including the known deafness gene MYO6. No pathogenic variants were found in MYO6, neither with sequence analysis of the coding region and splice sites nor with mRNA analysis. Subsequent candidate gene evaluation revealed CLIC5 as an excellent candidate gene. The orthologous mouse gene is mutated in the jitterbug mutant that exhibits progressive hearing impairment and vestibular dysfunction. Mutation analysis of CLIC5 revealed a homozygous nonsense mutation c.96T>A (p.(Cys32Ter)) that segregated with the hearing loss. Further analysis of CLIC5 in 213 arNSHI patients from mostly Dutch and Spanish origin did not reveal any additional pathogenic variants. CLIC5 mutations are thus not a common cause of arNSHI in these populations. The hearing loss in the present family had an onset in early childhood and progressed from mild to severe or even profound before the second decade. Impaired hearing is accompanied by vestibular areflexia and in one of the patients with mild renal dysfunction. Although we demonstrate that CLIC5 is expressed in many other human tissues, no additional symptoms were observed in these patients. In conclusion, our results show that CLIC5 is a novel arNSHI gene involved in progressive hearing impairment, vestibular and possibly mild renal dysfunction in a family of Turkish origin.
Similar content being viewed by others
Introduction
Hearing impairment is the most common sensory disorder worldwide and it is clinically and genetically very heterogeneous.1 Approximately 80% of early onset hereditary nonsyndromic hearing impairment inherits in an autosomal recessive pattern. Currently, 80 loci and 49 genes have been identified for autosomal recessive nonsyndromic hearing impairment (arNSHI), showing the great genetic heterogeneity (Hereditary Hearing Loss Homepage, http://www.hereditaryhearingloss.org/). This heterogeneity might well be explained by the complexity of the auditory system. Defects in a large variety of biological processes such as gene regulation, ion homeostasis and hair bundle morphogenesis can lead to hearing impairment.2
In the last decade, homozygosity mapping using genome-wide single nucleotide polymorphism (SNP) genotyping has been a powerful tool in the identification of arNSHI loci and genes.3 Lately, next generation sequencing technologies have revolutionized the genetics field and also led to the identification of novel arNSHI genes at fast pace.4 The most powerful evidence to assign a candidate gene as a novel deafness gene is to discover pathogenic variants in several families and not in controls. However, due to the large genetic heterogeneity of hearing impairment, this can be very difficult even with the current technologies. This is also evident from three recently identified arNSHI genes, PNPT1, SERPINB6 and TSPEAR, for which mutations have only been described in a single family each.5, 6, 7 Further evidence to assign a candidate gene as a deafness gene can come from animal models, especially mouse models with hearing loss. Several genes essential for hearing in humans were identified after they had already been demonstrated to be associated with deafness in mice.8
Using homozygosity mapping and candidate gene analysis, we identified a homozygous nonsense mutation in CLIC5 in a consanguineous Turkish family (W05-009). The orthologous mouse gene, Clic5, was described to be mutated in the jitterbug (jbg) mouse exhibiting congenital progressive hearing impairment and vestibular dysfunction due to progressive hair cell degeneration.9
Materials and methods
Subjects and clinical evaluations
This study was approved by the local medical ethics committee of the Radboud University Medical Center and Hospital Universitario Ramon y Cajal. Signed informed consent was obtained from the parents, as all patients are minors. In addition, a signed form was used to retrieve relevant data from other medical centers.
For the affected individuals of family W05-009, general physical examination was performed by a pediatrician. Blood and urine samples were analyzed to evaluate renal and thyroid function. Ear, nose and throat examination was executed to exclude other possible causes of hearing impairment like previous ear surgery and external ear deformities. A computed tomography scan of the temporal bone was performed in order to exclude possible anatomical causes of hearing loss. Pure tone audiometry was performed according to current standards to determine hearing thresholds at 0.25, 0.5, 1, 2, 4 and 8 kHz. To exclude conductive hearing loss, both air conduction and bone conduction thresholds were obtained. Classification of the hearing loss is in accordance with the GENDEAF guidelines (Hereditary Hearing Loss Homepage, http://www.hereditaryhearingloss.org/). In addition, otoacoustic emissions were measured in individual II.3. Vestibular function was evaluated by electronystagmography and rotatory tests.10 GraphPad Prism 5.00 (GraphPad, San Diego, CA, USA) was used to perform linear regression analysis to evaluate progression of the hearing impairment.
Three panels of arNSHI patients were analyzed for the involvement of CLIC5. GJB2 mutations or GJB6 deletions were excluded by routine analysis in most of these patients. The first panel consisted of 76 arNSHI index patients, mostly of Dutch origin, and these were selected based on the hearing loss phenotype. They presented either with a downsloping audiogram configuration and progression of hearing loss or early onset progressive hearing loss accompanied by vestibular areflexia or hyporeflexia. The second panel consisted of 69 unrelated arNSHI sibships of Spanish origin, which were not preselected based on type or severity of their hearing impairment. The third panel contained 18 arNSHI index patients of Spanish origin selected based on the hearing loss phenotype. In most of the cases, the hearing loss was postlingual (16 in childhood, at school age; two in the second decade of life) and progressive with a downsloping audiogram configuration.
Homozygosity mapping
Genomic DNA was isolated from peripheral blood lymphocytes by standard procedures. Individuals II.2 and II.3 from family W05-009 were genotyped using the Affymetrix mapping 250K NspI SNP array. All SNP array experiments were performed and analyzed according to the manufacturer's protocol (Affymetrix, Santa Clara, CA, USA). Genotype calling and calculation of the regions of homozygosity were performed with the Genotyping Console software (Affymetrix) with the default settings. The cosegregation of the genotypes for each previously reported arNSHI gene was visually evaluated.
Mutation analysis
Primers for amplification of exons and exon–intron boundaries of CLIC5 (NM_016929.4, CLIC5A and NM_001114086.1, CLIC5B), ESPN (NM_031475.2), MYO6 (NM_004999.3) and for mRNA analysis of MYO6 (NM_004999.3) were designed with ExonPrimer (http://www.ihg.gsf.de/ihg/ExonPrimer.html). Primer sequences and PCR conditions are provided in Supplementary Table S1. Amplification by PCR was performed on 40 ng of genomic DNA with Taq DNA polymerase (Roche, Mannheim, Germany). For MYO6 mRNA analysis, total RNA was isolated from Epstein–Barr virus-transformed lymphoblastoid cells of affected individual II.2 using the NucleoSpin RNA II kit (Machery Nagel, Düren, Germany) according to the manufacturer’s protocol. Subsequently, cDNA synthesis was performed with 1.5 μg RNA as starting material by using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA, USA), according to the manufacturer’s protocol. PCR reactions were performed on 2 μl cDNA with the Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA).
PCR fragments were purified with the use of NucleoFast 96 PCR plates (Clontech, Mountain View, CA, USA) or ExoI/FastAP (Fermentas, Vilnius, Lithuania) in accordance with the manufacturer’s protocol. Sequence analysis was performed with the ABI PRISM BigDye Terminator Cycle Sequencing V2.0 Ready Reaction kit and analyzed with the ABI PRISM 3730 DNA analyzer (Applied Biosystems Foster City, CA, USA). The presence of the CLIC5 c.96T>A transversion was investigated in 111 ethnically matched healthy controls. Exon 2 of CLIC5 was amplified and PCR products were purified as described above. Digestion of the PCR products with HpyCH4III (New England Biolabs, Ipswich, MA, USA) was performed in accordance with the manufacturer’s protocol, and restriction fragments were analyzed on gels containing 1.5% agarose and 1% low-melting agarose. The mutation removes a restriction site.
Nonsense-mediated mRNA decay evaluation
Epstein–Barr virus-transformed lymphoblastoid cell lines were established from heparin blood of individuals II.2 and II.3. Cells were grown with and without cycloheximide, a protein synthesis inhibitor, which prevents the nonsense-mediated mRNA decay (NMD) process as described previously.11 Total RNA was isolated as described above. cDNA synthesis was performed with 3 μg RNA as starting material by using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories), according to the manufacturer’s protocol. For the quantitative PCR (qPCR), specific primers (Supplementary Table S1) were designed with Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) and reference sequence NM_016929.4. PCRs were performed with the Applied Biosystem Fast 7900 System in accordance with the manufacturer’s protocol. The human beta glucuronidase gene (GUSB (MIM 611499)) was employed as an internal control. PCR mixtures were prepared with the Power Syber Green Master Mix (Applied Biosystems) in accordance with the manufacturer’s protocol. Temperatures and reaction times for PCR were as follows: 10 min at 95 °C, followed by 40 cycles of 15 s at 95 °C and 30 s at 60 °C. All reactions were performed in duplicate. Relative gene expression levels were determined with the ΔΔCt method as described previously.11
CLIC5 expression profile
RNA derived from adult heart, retina, brain and kidney was purchased from Clontech (Mountain View, CA, USA). RNA derived from adult skeletal muscle, liver, duodenum, testis, spleen, thymus and placenta was purchased from Stratagene (La Jolla, CA, USA) and bone marrow from Bio-chain (Newark, CA, USA). RNA derived from fetal brain, colon, kidney, stomach, spleen, heart, skeletal muscle, lung and thymus was purchased from Stratagene. In addition, RNA was isolated from adult lung, fetal cochlea and fetal liver as described previously.12 The inner ear was derived from a fetus at 8 weeks of gestation and the other fetal tissues from fetuses at 20–21 weeks of gestation. cDNA synthesis, primer design (Supplementary Table S1) and qPCR analysis were performed as described above. The forward primer was located on the boundary of exons 2 and 3, and the reverse primer in exon 3. This enabled detection of all CLIC5 isoforms affected by the nonsense mutation that is located in exon 2. Relative gene expression levels were determined with the comparative ΔCt method as described previously.13
Results
Hearing loss and vestibular dysfunction in family W05-009
Ear, nose and throat examination and computed tomography scanning did not reveal an apparent cause of hearing impairment in the two affected children of family W05-009. The parents have normal hearing and are first cousins of Turkish ancestry (Figure 1a). The patients presented with an early onset sensorineural hearing loss since the bone conduction thresholds did not differ from the air conduction thresholds. The hearing impairment probably was not congenital as individual II.2 passed a behavioral reflex audiometry test which is performed between 7 and 9 months of age (Ewing test) and individual II.3 had normal hearing at the age of 3 months during brainstem-evoked response audiometry. The hearing loss started mildly, affecting the mid and high frequencies mostly. It progressed to a severe-to-profound hearing loss with a gently-to-steeply downsloping audiogram configuration as is shown in Figure 1b. Longitudinal linear regression analysis indicated significant progression in all frequencies (Figure 1c). Otoacoustic emissions were not present in individual II.3 at the age of 4 months. Individual II.2 received a cochlear implant at the age of 11 years. Five years post implantation, the speech recognition scores were 88% (using the standard Dutch phonetically balanced consonant-vocal-consonant word lists).14 Initial motor milestones were reported to be normal, but later in life balance problems did occur, for example, difficulties with walking in the dark and cycling. Vestibular areflexia was found in both individuals during the rotatory test at the age of 16 years (II.2) and 11 years (II.3).
(a) Pedigree of family W05-009 and segregation of the CLIC5 c.96T>A variant. (b) Longitudinal binaural mean air-conduction pure tone thresholds are shown of affected members of family W05-009. Age (years) is shown with a symbol key. The p95 line, matched for age and sex, indicates that 95% of the population has thresholds lower than these. (c) Regression analysis of longitudinal binaural mean air conduction threshold data for each frequency separately. Circles indicate individual II.2, squares indicate individual II.3. Annual threshold deterioration is shown behind the symbol key for each frequency.
General physical examination showed that both children had a normal height and weight. No other abnormalities besides the hearing impairment were noted in individual II.2. On history, there were no signs of any other abnormalities in individual II.3. Thyroid function was also normal in individuals II.2 and II.3. Renal function was normal in II.2. However, in individual II.3, a blood pressure of 127/73 to 129/88 mm Hg (90% value for this age and length: 120/76 mm Hg) was measured and an elevated albumin/creatinine ratio was detected at the last two visits in the outpatient clinic (9.2 and 3.8 mg/mmol, respectively, normal value <2.5 mg/mmol) during urine analysis at the age of 11 years. The glomerular filtration rate, calculated with the Schwartz equation, was normal with a value of 114 ml/min per 1.73 m2 (normal>90 ml/min per 1.73 m2). As the elevated blood pressure and albumin/creatinine ratio were seen in repeated measurements, these might be the first signs of a nephropathy. However, no other more invasive studies were performed, since the renal function was overall only mildly affected. Subsequent renal function follow-up of patient II.3 in the future will be needed to determine whether it is indeed a nephropathy.
Homozygosity mapping confines a critical region on chromosome 6p21.1-q15
To identify genes for arNSHI, homozygosity mapping was performed in some large series of familial and isolated arNSHI patients living in the Netherlands. In family W05-009, homozygosity mapping revealed a homozygous region of 47.4 Mb on chromosome 6p21.1-q15 (Figure 2a). This was the largest homozygous region shared by the two affected individuals. This region contains the known deafness gene MYO6. There were 15 other shared homozygous regions larger than 1 Mb (Supplementary Table S2), and in one of them ESPN is located, which is another gene known for arNSHI. Mutation analysis was performed for all exons and exon–intron boundaries of ESPN and MYO6, which revealed no putative causative variants. As MYO6 was located in the largest shared homozygous region, MYO6 mRNA was analyzed by RT-PCR. No PCR products of an aberrant size were indentified and also sequence analysis of the PCR products did not reveal indications for aberrant splicing. To exclude compound heterozygous or allozygous mutations in the other known arNSHI genes, cosegregation of the genotypes was visually evaluated for each gene. PNPT1, ILDR1, RDX, TECTA, OTOGL, PTPRQ and OTOA were present within the genotype-shared regions. OTOA could be excluded by short tandem repeat marker analysis in the family. No putative pathogenic variants were identified in the other genes by sequence analysis of the exons and exon–intron boundaries.
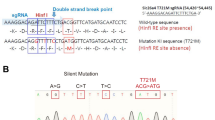
(a) Schematic overview of the chromosomal region 6q21.1-q15 showing the homozygous regions (black bars) identified in the affected individuals of family W05-009. The homozygous region of individual II.2 delimits the region to 47.4 Mb. Mb positions and chromosome bands are according to the UCSC genome browser (GRCh37). (b) Partial sequences are shown of CLIC5 exon 2 from an affected member, a parent and an unaffected sib of family W05-009. The predicted amino-acid changes and the surrounding amino acids are indicated above the sequence. Mutated nucleotides are marked by an arrowhead. As reference sequence NM_016929.4 was employed.
A nonsense mutation in CLIC5 causes arNSHI
As no pathogenic variants could be identified in MYO6 or ESPN, we hypothesized that another gene would be underlying the hearing loss in this family. Therefore, we continued with the evaluation of the 247 annotated genes in the largest homozygous region, 6p21.1-q15 (http://www.genome.ucsc.edu/, RefSeq genes, hg19; assigned DFNB102). For three of the genes, mutations in the orthologous mouse gene cause deafness. Slc17a5 (MGI:1924105)15 and Bmp5 (MGI:88181) are associated with mixed and conductive hearing loss in the mouse, respectively. The third gene, Clic5, was described to underlie progressive sensorineural hearing loss and vestibular dysfunction in the jbg mouse mutant.9 Therefore, CLIC5 represented an excellent candidate gene.
Mutation analysis of CLIC5 revealed a homozygous nonsense variant c.96T>A (p.(Cys32Ter)) (Figure 2b), which segregated with the hearing impairment in family W05-009 (Figure 1a). This variant was not present in 222 Turkish control alleles, the Exome Variant Server (http://www.evs.gs.washington.edu/EVS/) and the Nijmegen in-house exome database (1302 exomes). This variant was submitted to the Leiden Open Variant Database (http://www.databases.lovd.nl/shared/variants/0000022995).
The homozygous CLIC5 c.96C>T mutation is predicted to result in NMD, as it creates a premature stop codon (p.(Cys32Ter)) more than 54 bp upstream of the 3'-most intron.16 However, we could not confirm NMD. The relative CLIC5 mRNA expression is comparable in cycloheximide-treated and -untreated patient Epstein–Barr virus-cell lines (157.24 vs 157.89%, respectively) and higher—albeit not significantly—than in controls (set at 100%) as shown in Supplementary Figure 1.
For identification of other families with CLIC5 mutations, sequence analysis of the coding region and splice sites of CLIC5 was performed in 76 arNSHI index patients, mostly of Dutch origin, and 18 Spanish arNSHI index patients. These patients were preselected based on the hearing loss and vestibular phenotype as described in the Materials and methods section. No putatively causative variants were identified in these patients. In a parallel approach, compatibility with linkage to DFNB102 was investigated in a panel of 69 unrelated arNSHI sibships of Spanish origin. These were genotyped for short tandem repeat-markers D6S459, D6S1920, D6S1632 and D6S1638, which flank CLIC5. Haplotype analysis revealed compatibility with linkage of the disease locus to these markers in 18 sibships. Sequence analysis of CLIC5 in the index cases of these sibships did not reveal any putative pathogenic variants. Finally, data of whole-exome sequence analysis of 50 arNSHI patients, mainly of Dutch origin, were evaluated for the presence of variants in CLIC5. These patients were not preselected based on the type or severity of the hearing loss. No putatively pathogenic variants were identified in this cohort.
CLIC5 is expressed in the human inner ear
The expression of CLIC5 was studied via qPCR in human fetal inner ear and compared to that in an additional 13 adult- and 10 fetal-stage human tissues (Figure 3). As this was performed in two separate experiments for adult and fetal tissues, fetal inner ear was included in both for comparison. The tissues were not derived from fetuses of the same gestational stage; therefore, a direct comparison of the transcript levels cannot be made. In fetal tissues, CLIC5 transcript levels were highest in skeletal muscle, kidney, spleen, heart and colon. A lower expression level was found in fetal brain, thymus, lung and stomach. Expression in the fetal inner ear was 26-fold higher than in fetal liver in which the expression level was the lowest. In adult tissues, the highest expression levels were found in heart, lung, skeletal muscle and kidney and there was a gradual decrease through retina, spleen, brain, placenta, duodenum and thymus. Finally, the lowest expression levels were found in bone marrow, testis and liver. We can conclude that CLIC5 is widely expressed in both fetal and adult human tissues.
CLIC5 expression profile in human tissues. Relative CLIC5 mRNA levels as determined by qPCR in human fetal (a) and adult (b) tissues. The relative expression values were determined by using the comparative delta Ct method.13 The expression levels are relative to those in liver, which showed the lowest CLIC5 expression of all the tissues that were tested.
Discussion
We present a homozygous nonsense mutation, c.96T>A (p.(Cys32Ter)) in CLIC5 (DFNB102) as a cause of sensorineural hearing impairment accompanied by vestibular dysfunction. This nonsense variant will lead to an early truncation of the protein. Screening a series of arNSHI index patients did not reveal additional causative variants, suggesting that mutations in CLIC5 are not a common cause of arNSHI neither in the Netherlands nor in Spain.
Initially, the hearing loss in the affected individuals of family W05-009 was mild, mainly affecting the mid and high frequencies, and then it progressed to severe-to-profound hearing loss. They also showed vestibular areflexia in the second decade of life. The combination of progression to profound hearing loss and complete vestibular dysfunction in the patients of family W05-009 resembles the phenotype of the jbg mutants. Auditory-evoked brainstem responses at 1–5 months of age in jbg-mutant mice, which are null for Clic5, were 40–50 dB above those of wild-type mice. By 7 months of age, their hearing loss had progressed to complete deafness due to progressive hair bundle degeneration and a reduced density of spiral ganglion cells.9 The vestibular hair cells of jbg mice also showed a progressive degeneration. In the crista ampullaris, the number of vestibular hair cells was reduced at 3 months and hair cells were nearly absent at 12 months of age.9
Besides inner ear dysfunction, the phenotype in humans and mice with Clic5 mutations may well overlap with respect to the renal phenotype. The jbg mice have abnormalities in the foot processes of the kidney podocytes leading to proteinuria.17, 18 The elevated albumin/creatinine ratio and pre-hypertension in affected individual II.3 indicate mild renal dysfunction and may well be the first signs of a nephropathy. Therefore, follow-up of renal function is indicated for individual II.3 but also for his hearing-impaired sister.
In addition to the inner ear and kidney phenotypes, the jbg-mutant mice also exhibit emphysema-like lung pathology, hyperactivity and resistance to diet-induced obesity.19 Characteristics of lung emphysema and hyperactivity were excluded in family W05-009 by history taking. The affected individuals had normal height and weight. No other gross abnormalities in the organs of the jbg mutants were detected and no other complaints from the affected siblings were reported so far. The lack of symptoms in organs with relatively high CLIC5 expression (Figure 3) points toward redundancy for CLIC5 function there.
CLIC5 belongs to a family of chloride intracellular channel proteins, but its protein structure differs from other typical ion-channel proteins. CLICs (CLIC1-6) show no sequence homology to any known channel family, but significant homology to glutathione S-transferases in the core region.20 Moreover, some of the CLIC proteins, including CLIC5, are not only integral membrane proteins, but are also found as soluble proteins in the cytoplasm.21 In the inner ear, CLIC5 localizes to stereocilia of both the cochlear and vestibular hair cells and on the surface of Kolliker’s organ during cochlea development in mice.9 Specifically, CLIC5 is predominantly present at the base of stereocilia and, to lesser extent, in the cell bodies of hair cells in the region surrounding the cuticular plate.9 CLIC5 was initially identified in placental microvilli as a component of a multimeric complex consisting of several known cytoskeletal proteins, including actin and ezrin.22 Although the function of CLIC5 is still elusive, there are several facts that support its role in stereocilia integrity. Firstly, radixin immunostaining is reduced in the hair bundle of jbg mice where it colocalizes with Clic5.9 This suggests that Clic5 is needed for proper radixin activity, so when interacting with the erzin, radixin and moesin complex, Clic5 may stabilize connections between the plasma membrane and the filamentous actin core.9 Secondly, Clic5 functions as an adapter between the plasma membrane of podocytes and the actin cytoskeleton by facilitating the interaction between ezrin and podocalyxin.17, 18, 23 Thirdly, a recent study proposes that Clic5 functions as part of a multiprotein linker complex in companion with radixin, erzin and taperin.24 Protein tyrosine phosphatase receptor Q (Ptprq), which is mislocalized as radixin in the jbg mice, and Myosin VI, key regulator of the proper localization of Ptprq,25 might well participate in this complex too. Radixin,26 Ptprq25, 27 Myosin VI28 and Clic5-deficient mice9 show loss of stereocilia at the bundle vertex and fusion of stereocilia in postnatal stage. This suggests that this multiprotein complex is essential for stable membrane-cytoskeletal attachments at the stereocilia base.
In conclusion, we show that a homozygous nonsense mutation in CLIC5 (p.(Cys32Ter)) underlies the progressive hearing impairment, vestibular and possibly mild renal dysfunction in a family of Turkish origin. CLIC5 mutations do not seem to be a common cause of arNSHI in the Dutch and Spanish populations. Further screening of CLIC5 in other populations will provide important information about the frequency of CLIC5 mutations and may aid to further define the phenotype associated with CLIC5 mutations.
References
Mehl AL, Thomson V : The Colorado newborn hearing screening project, 1992-1999: on the threshold of effective population-based universal newborn hearing screening. Pediatrics 2002; 109: E7.
Dror AA, Avraham KB : Hearing loss: mechanisms revealed by genetics and cell biology. Annu Rev Genet 2009; 43: 411–437.
Schraders M, Lee K, Oostrik J et al: Homozygosity mapping reveals mutations of GRXCR1 as a cause of autosomal-recessive nonsyndromic hearing impairment. Am J Hum Genet 2010; 86: 138–147.
Shearer AE, Smith RJ : Genetics: advances in genetic testing for deafness. Curr Opin Pediatr 2012; 24: 679–686.
Delmaghani S, Aghaie A, Michalski N, Bonnet C, Weil D, Petit C : Defect in the gene encoding the EAR/EPTP domain-containing protein TSPEAR causes DFNB98 profound deafness. Hum Mol Genet 2012; 21: 3835–3844.
Sirmaci A, Erbek S, Price J et al: A truncating mutation in SERPINB6 is associated with autosomal-recessive nonsyndromic sensorineural hearing loss. Am J Hum Genet 2010; 86: 797–804.
von Ameln S, Wang G, Boulouiz R et al: A mutation in PNPT1, encoding mitochondrial-RNA-import protein PNPase, causes hereditary hearing loss. Am J Hum Genet 2012; 91: 919–927.
Friedman LM, Dror AA, Avraham KB : Mouse models to study inner ear development and hereditary hearing loss. Int J Dev Biol 2007; 51: 609–631.
Gagnon LH, Longo-Guess CM, Berryman M et al: The chloride intracellular channel protein CLIC5 is expressed at high levels in hair cell stereocilia and is essential for normal inner ear function. J Neurosci 2006; 26: 10188–10198.
Theunissen EJ, Huygen PL, Folgering HT : Vestibular hyperreactivity and hyperventilation. Clin Otolaryngol Allied Sci 1986; 11: 161–169.
Livak KJ, Schmittgen TD : Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402–408.
Luijendijk MW, van de Pol TJ, van Duijnhoven G et al: Cloning, characterization, and mRNA expression analysis of novel human fetal cochlear cDNAs. Genomics 2003; 82: 480–490.
Pfaffl MW : A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29: e45.
Bosman AJ SG : Woordenlijst Voor Spraakaudiometrie (Compact Disc) Gouda. Electro Medical Instruments bv & Veenhuis Medical Audio bv: The Netherlands, 1992.
Prolo LM, Vogel H, Reimer RJ : The lysosomal sialic acid transporter sialin is required for normal CNS myelination. J Neurosci 2009; 29: 15355–15365.
Nagy E, Maquat LE : A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci 1998; 23: 198–199.
Pierchala BA, Munoz MR, Tsui CC : Proteomic analysis of the slit diaphragm complex: CLIC5 is a protein critical for podocyte morphology and function. Kidney Int 2010; 78: 868–882.
Wegner B, Al-Momany A, Kulak SC et al: CLIC5A, a component of the ezrin-podocalyxin complex in glomeruli, is a determinant of podocyte integrity. Am J Physiol Renal Physiol 2010; 298: F1492–F1503.
Bradford EM, Miller ML, Prasad V et al: CLIC5 mutant mice are resistant to diet-induced obesity and exhibit gastric hemorrhaging and increased susceptibility to torpor. Am J Physiol Regul Integr Comp Physiol 2010; 298: R1531–R1542.
Dulhunty A, Gage P, Curtis S, Chelvanayagam G, Board P : The glutathione transferase structural family includes a nuclear chloride channel and a ryanodine receptor calcium release channel modulator. J Biol Chem 2001; 276: 3319–3323.
Ashley RH : Challenging accepted ion channel biology: p64 and the CLIC family of putative intracellular anion channel proteins (Review). Mol Membr Biol 2003; 20: 1–11.
Berryman M, Bretscher A : Identification of a novel member of the chloride intracellular channel gene family (CLIC5) that associates with the actin cytoskeleton of placental microvilli. Mol Biol Cell 2000; 11: 1509–1521.
Jiang L, Phang JM, Yu J et al: CLIC proteins, ezrin, radixin, moesin and the coupling of membranes to the actin cytoskeleton: a smoking gun? Biochim Biophys Acta 2014; 1838: 643–657.
Salles FT, Andrade LR, Tanda S et al: CLIC5 stabilizes membrane-actin filament linkages at the base of hair cell stereocilia in a molecular complex with radixin, taperin, and myosin VI. Cytoskeleton (Hoboken) 2013; 71: 61–78.
Sakaguchi H, Tokita J, Naoz M, Bowen-Pope D, Gov NS, Kachar B : Dynamic compartmentalization of protein tyrosine phosphatase receptor Q at the proximal end of stereocilia: implication of myosin VI-based transport. Cell Motil Cytoskeleton 2008; 65: 528–538.
Kitajiri S, Fukumoto K, Hata M et al: Radixin deficiency causes deafness associated with progressive degeneration of cochlear stereocilia. J Cell Biol 2004; 166: 559–570.
Goodyear RJ, Legan PK, Wright MB et al: A receptor-like inositol lipid phosphatase is required for the maturation of developing cochlear hair bundles. J Neurosci 2003; 23: 9208–9219.
Self T, Sobe T, Copeland NG, Jenkins NA, Avraham KB, Steel KP : Role of myosin VI in the differentiation of cochlear hair cells. Dev Biol 1999; 214: 331–341.
Acknowledgements
We are grateful to the family for their participation in this study. We thank Saskia van der Velde-Visser, Marloes Steehouwer and Irene Janssen for excellent technical assistance, Ersan Kalay for providing Turkish control DNA samples, Arjan de Brouwer for his contribution to the NMD analysis and Rob Collin for SNP-array analysis. This work was financially supported by grants from the Heinsius Houbolt Foundation (to HK), The Oticon Foundation (09-3742, to HK), ZorgOnderzoek Nederland/Medische Wetenschappen (40-00812-98-09047, to HK, 90700388 to RJEP and 016.136.088 to MS), the Netherlands Genomics Initiative (40-41009-98-9073, to MS) and Instituto de Salud Carlos III (FIS PI11/00612, to IdC).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Seco, C., Oonk, A., Domínguez-Ruiz, M. et al. Progressive hearing loss and vestibular dysfunction caused by a homozygous nonsense mutation in CLIC5. Eur J Hum Genet 23, 189–194 (2015). https://doi.org/10.1038/ejhg.2014.83
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2014.83
This article is cited by
-
ARNSHL gene identification: past, present and future
Molecular Genetics and Genomics (2022)
-
Identification of autosomal recessive nonsyndromic hearing impairment genes through the study of consanguineous and non-consanguineous families: past, present, and future
Human Genetics (2022)
-
Inherent flexibility of CLIC6 revealed by crystallographic and solution studies
Scientific Reports (2018)
-
Genetic contribution to vestibular diseases
Journal of Neurology (2018)
-
The diagnostic yield of whole-exome sequencing targeting a gene panel for hearing impairment in The Netherlands
European Journal of Human Genetics (2017)