Abstract
The imprinted region on chromosome 14q32 harbors several maternally or paternally expressed genes as well as two DMRs (differentially methylated regions), the IG-DMR and the MEG3-DMR, which both act as imprinting control centers. Genetic aberrations affecting the imprinted gene cluster in 14q32 result in distinct phenotypes, known as maternal or paternal uniparental disomy 14 phenotypes (upd(14)mat, upd(14)pat). In both syndromes, three types of molecular alterations have been reported: uniparental disomy 14, deletions and epimutations. In contrast to uniparental disomy and epimutations, deletions affecting regulatory elements in 14q32 are associated with a high-recurrence risk. Based on two single deletion cases a functional hierarchy of the IG-DMR as a regulator for the methylation of the MEG3-DMR has been proposed. We have identified two novel deletions of maternal origin spanning the MEG3-DMR, but not the IG-DMR in patients with upd(14)pat syndrome, one de novo deletion of 165 kb and another deletion of 5.8 kb in two siblings. The 5.8 kb deletion was inherited from the phenotypically normal mother, who carries the deletion in a mosaic state on her paternal chromosome 14. The methylation at both DMRs was investigated by quantitative next generation bisulfite sequencing and revealed normal methylation patterns at the IG-DMR in all patients with the exception of certain CpG dinucleotides. Thus, we could confirm that deletions of the MEG3-DMR does not generally influence the methylation pattern of the IG-DMR, which strengthens the hypothesis of a hierarchical structure and distinct functional properties of the two DMRs.
Similar content being viewed by others
Introduction
The human chromosomal region 14q32 carries a cluster of imprinted genes, which includes the paternally expressed genes delta-like 1 homolog, Drosophila (DLK1) and RTL1 (Retroposon-like 1, PEG11) as well as the maternally expressed genes MEG3 (alias GTL2), RTL1as, MEG8, numerous C/D box small nucleolar (sno)RNAs and microRNAs.1, 2, 3, 4 In addition, the 14q32-imprinted region harbors two differentially methylated regions (DMRs), the germline-derived primary intergenic DMR between DLK1 and MEG3 (IG-DMR) and the postfertilization-derived secondary MEG3-DMR, which are methylated on the paternal and unmethylated on the maternal chromosome 14 in most cells (see Figure 1).5, 6, 7, 8 Genetic and epigenetic alterations affecting the imprinted gene cluster in 14q32 result in two different phenotypes currently known as maternal or paternal uniparental disomy 14 phenotypes (upd(14)mat, upd(14)pat). The upd(14)pat syndrome is characterized by polyhydramnios, a small bell-shaped thorax, facial dysmorphisms, abdominal wall defects, distal arthrogryposis and mental retardation,9, 10 whereas the upd(14)mat syndrome is characterized by pre and postnatal growth retardation, neonatal hypotonia, feeding problems and precocious puberty.11, 12, 13 In both syndromes, three types of molecular alterations have been reported: uniparental disomy, deletions and epimutations.7, 14, 15, 16, 17, 18, 19 In contrast to uniparental disomy and epimutations, deletions affecting regulatory elements in 14q32 on the maternal or the paternal allele are associated with a high-recurrence risk.7, 19 It has been shown that both the MEG3- and the IG-DMR function as imprinting control centers in the placenta and the body. A functional hierarchy of the IG-DMR as a regulator for the methylation pattern of the MEG3-DMR has been postulated based on a patient with a maternally inherited deletion of 8.6 kb affecting the IG-DMR but not the MEG3 promoter,19 which was associated with an epimutation (hypermethylation) of the MEG3-DMR and a phenotype in body and placenta. In contrast, in another patient in the same study with a deletion of 4.3 kb on the maternal allele affecting the MEG3-DMR but not the IG-DMR, this deletion was associated with normal methylation at the IG-DMR (Figure 1). In this patient the placenta showed normal appearance. In both patients the deletion results in a typical upd(14)pat syndrome.
Schematic overview of the imprinted region on chromosome 14q32. The scheme depicts the deletions described here (patients 1, 2 and 3) and in a previous report by Kagami et al.19 in relation to the imprinted genes in the region 14q32. The MS-MLPA probes of the region are indicated by black and gray lines. Black lines represent MLPA probes for dosage; gray lines represent MLPA probes for dosage and methylation. Blue, paternally expressed genes; red, maternally expressed genes; gray, DMRs (differentially methylated regions). Not drawn to scale.
Here we report on the identification and molecular characterization of two additional cases with deletions on chromosome 14q32 affecting the MEG3-DMR but not the IG-DMR: one single patient with a maternal de novo microdeletion and two siblings with a microdeletion inherited from their mother who carries this deletion in a mosaic state on her paternal chromosome 14. To examine the aforementioned hypothesis of a hierarchical interaction and the distinct functional properties of the two DMRs more precisely we performed quantitative methylation analyses in our patients and the patient with the isolated 4.3 kb MEG3-DMR deletion published by Kagami et al19 by next generation bisulfite sequencing of both DMRs.
Materials and methods
Subjects and samples
Three individuals with clinical features suggestive for upd(14)pat syndrome were referred to the departments of Human Genetics in Lübeck (patient 1) and Aachen (patients 2 and 3). The families agreed to participate in the molecular studies. The studies were approved by the ethics committees of the participating institutions. A DNA sample from the patient with the 4.3 kb MEG3 deletion published before19 was kindly provided by Andrew Green.
DNA extraction
DNA was extracted from peripheral lymphocytes using the Flexigene Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocols, or by a simple salting-out procedure.
Methylation-specific PCR (MS-PCR)
Genomic DNA was purified from whole blood and treated with sodium bisulfite according to standard methods. MS-PCR analysis of the MEG3-DMR was carried out as described by Dietz et al.20 Touchdown PCR was used as follows: annealing temperatures were 5 cycles at 70 °C for 30 s, 5 cycles at 65 °C for 30 s and 30 cycles for 60 °C.
Methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA)
Gene dosage of the imprinted DLK1/MEG3 locus and methylation of the MEG3 promoter region on chromosome 14q32 were analyzed by MS-MLPA using the SALSA MLPA KIT ME032-X1 kit (MRC Holland, Amsterdam, Netherlands). This newly established MS-MLPA kit contains 21 probes for gene dosage analysis of the chromosomal region 14q32 and three methylation sensitive probes for the MEG3 promoter region. Hybridization, ligation and PCR reactions were carried out according to the manufacturer’s instructions. Amplification products were analyzed by capillary electrophoresis using the ABI3100 capillar sequencer. Data analysis was carried out using the Gene Marker Software (Softgenetics, State College, PA, USA).
Microarray analysis
For genome-wide copy number analysis the Affymetrix human SNP array 6.0 (Affymetrix, Santa Clara, CA, USA) was carried out following the manufacturer’s protocol. Data analysis was performed with the Genotyping Console and Chas software (Affymetrix).
Long-range PCR
To determine the exact size of the deletions and to identify the breakpoints on the DNA sequence level a long-range PCR was carried out using the Expand long-range dNTPack (Roche, Mannheim, Germany). Primers were generated spanning the sites of MLPA probes which showed a normal dosage in the siblings (patients 2 and 3) or spanning SNP or copy number probes of the array in patient 1 (for primer sequences see Supplementary Table 1). PCR was performed according to the manufacturer’s manual with a magnesium-chloride (MgCl2) containing buffer, a final primer concentration of 0.6 μ M, with or without 6% DMSO and a DNA input of about 200 ng. The PCR program was carried out as described in the manual with an elongation time of 9 min and 28 cycles or 8 min and 20 cycles respectively in the third stage. The PCR-product derived from the allele harboring the deletion was either gel purified using the QiaQuick gel extraction Kit (Qiagen) or by ExoSAP-It (USB, Santa Clara, CA, USA) as described in the manufacturers’ protocols. Sequencing was carried out with the BigDye Terminator v1.1 Cycle Sequencing Kit on an ABI3100 automatic capillary genetic analyzer (Applied Biosystems, Darmstadt, Germany). Sequence analysis was performed with Sequencing Analysis (Applied Biosystems) and Geneious (Biomatters, Auckland, New Zealand).
Quantitative PCR (qPCR)
A real-time PCR assay for the promoter region of MEG3 was performed as previously described by Buiting et al.16
Multilocus methylation-specific single nucleotide primer extension (MS-SNuPE)
To confirm the altered methylation pattern at the MEG3 locus, MS-SNuPE was performed as previously described by Begemann et al.21
DNA methylation analysis of the MEG3-DMR and IG-DMR by next generation bisulfite sequencing
Bisulfite treatment was performed using the EZ DNA Methylation-Gold Kit (Zymo Research Europe, Freiberg, Germany) according to the manufacturer’s instructions. For each individual, bisulfite amplicon libraries were generated and sample-specific barcode sequences were added. The amplicons were purified, diluted and clonally amplified in an emulsion PCR before sequencing on the Roche/454 GS junior system (Branford, CT, USA). A detailed description has been published previously.22 Primer sequences are listed in Supplementary Table 1. For subsequent data analysis the Amplikyzer software was used (https://bitbucket.org/ svenrahmann/amplikyzer/downloads). A minimum of 1200 reads for each sample was obtained. Only reads with a conversion rate over 95% were considered.
Results
Clinical findings
Patient 1
The patient was born to healthy non-consanguineous parents after 32+2 weeks of gestation. The pregnancy was complicated by polyhydramnion but placentomegaly was not reported. Measurements at birth were 2.6 kg (+1.6 SD), 47 cm (+1.4 SD) and head circumference 32 cm (+1.2 SD). The patient showed a hypoplastic thorax with ‘coat hanger’-like ribs, retromicrognathia and muscular hypotonia. He had feeding difficulties and needed assisted ventilation until the age of 3 months. At this time a tracheostoma was implanted which remained until the age of 11 months. Feeding difficulties with recurrent aspirations necessitated percutaneous endoscopic gastrostomy feeding until the age of 17 months when he got a MicKey Button. At the age of 3 y and 4 months oral feeding was still very difficult. He could walk without support. He spoke few words, comprehension was very limited. His height was 93 cm (−2 SD), weight 14,2 kg (mean), OFC 50,5 cm (mean), respectively. He still shows muscular hypotonia.
At the age of 15 months his height was 76 cm (−1.2 SD), weight 10.5 kg (Mean) and head circumference 49.3 cm (+0.4 SD), respectively. He still showed muscular hypotonia and generalized developmental delay. At the age of 27 months he was able to walk a few steps with support. He only spoke a few words. Comprehension was not age-appropriate.
Patients 2 and 3
The two children were born as the second and third child of consanguineous healthy Turkish parents (Figure 2a). The father’s height is 176 cm, the motheŕs height is 167 cm. Both parents successfully finished secondary education. The first pregnancy of the family ended with a preterm birth at 18 weeks, further information was not available. Apart from that, the family history was unremarkable. The oldest son (II-2) is healthy.
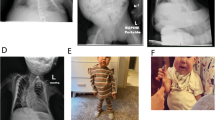
(a) Pedigree of patients 2 (II.3) and 3 (II.4) with upd(14)pat-syndrome phenotype. (b) X-rays of patients 2 and 3 showing the small, bell-shaped thorax with the characteristic ‘coat hanger’-like appearance of the ribs. (c) Patients 2 and 3. Patient 2 is shown at the age of 5, 6 or12 y and his less severe affected sister at 4 and 10 months.
Both younger children (patients 2 and 3, II-3 and II-4 in the pedigree) were delivered at 35 weeks of gestation because of polyhydramnion and preterm labor. Information about the placenta was not available.
Detailed clinical data of the siblings are listed in Table 1. The boy (II-3, patient 2) was delivered by Cesarean section because of pathological CTG records and asphyxia. Birth measurements were within the normal range (3125 g (1.5 SD), 46 cm (mean), 35 cm head circumference (2.1 SD)). The 1–5 and 10 min Apgar-scores were 1/4/4, marked muscular hypotonia was detected. Intubation and assisted ventilation were carried out for 16 days. Temporary occurring neonatal seizures were treated by Phenobarbital. His ventilation stabilized but he required oxygen administration until the age of 4 y. Laryngopharyngeal dysfunction with recurrent aspirations necessitated percutaneous endoscopic gastrostomy feeding for the first few years.
The girl (II-4, patient 3) was delivered vaginally with normal birth measurements (2800 g (1.1 SD), 45 cm (−0.1 SD), 32.5 cm head circumference (+0.7 SD)). By 1–5z and 10 min Apgar-scores of 5/2/5 and muscular hypotonia, she needed assisted ventilation by CPAP for the first 42 h but then no further ventilator support. She started sucking briefly after birth. In general the clinical course was milder in the girl than in her older brother.
Both children had the typical bell-shaped thorax with the ‘coat hanger’ appearance of the ribs on x-rays (Figure 2b) and the dysmorphic facial features such as short-palpebral fissures, anteverted nares, broad-nasal bridge, protruding philtrum and retromicrognathia. They showed frontal bossing with temple narrowing and full cheeks. The ears were deep set, the neck short and webbed (Figure 2c).
Both children, the boy more than the girl, had bilateral talipes calcaneovalgus and limited joint mobility especially in the wrist and fingers. Bilateral inguinal hernia repair was carried out in the boy at the age of 2 months, whereas his sister showed rectus diastasis and a small umbilical hernia.
The clinical findings of the boy improved over time. At the age of 5, 6 or 12 y (Figure 2c), he was able to walk without help and communicated in two word sentences. He needed no further medical support for ventilation or feeding. Formal intelligence testing had not been performed.
Molecular genetic findings
Methylation analysis in patient 1 performed by MS-PCR for the MEG3 promoter region revealed the presence of the methylated paternal allele only, indicating that the patient has upd(14)pat, an imprinting defect or a deletion affecting the MEG3 locus. Microsatellite analysis did not provide any evidence for uniparental disomy. Approximately 165 kb deletion on chromosome 14q32 was detected by Affymetrix genome-wide human SNP array 6.0 analysis (Santa Clara, CA, USA). The deletion includes MEG3, RTL1, MEG8 and most of the miRNA genes, but not the IG-DMR. MS-MLPA was used to confirm the deletion and to determine that the deletion affects the maternal allele (Figure 3a). MS-MLPA analysis in the mother’s DNA showed a normal result, indicating that the deletion in the patient occurred de novo (data not shown). Long-range PCR was used to amplify the deletion junction fragment and to further characterize the deletion size and breakpoints on the DNA sequence level (165.153 kb, hg19: chr14:g.101285913_101451066del see Figures 1 and 3b). Sequence comparison with a reference sequence (hg19) revealed a 6 bp insertion (TTCCCC) between the proximal and distal breakpoints (Figure 3b).
Results of the (a) MS-MLPA analysis and (b, c) the Sanger sequencing of the junction fragments obtained by long-range PCR for breakpoint identification. (a) The MS-MLPA for patient 1 showed a reduced dosage of about 50% for the nine probes encompassing MEG3, RTL1 and part of the miRNA cluster confirming the presence of a heterozygous deletion (plot and table). The probes for DLK1 and (the two probes) next to the IG-DMR displayed a normal dosage (around 1.0). A strong hypermethylation was demonstrated by the methylation-specific probes for the MEG3-DMR indicating that the deletion occurred on the unmethylated maternal allele (lower part of the table; normal methylation value should be ∼0.5). Results for the patients’ mother were normal (not shown), so that the deletion seemed to have occurred de novo. For patients 2 and 3 the dosage of three MLPA probes for the 5′ end of MEG3 are reduced by ∼50% indicating the presence of a heterozygous deletion (plot and table). Dosage reduction in the mother was ∼30%, pointing to a mosaic state of the deletion. Results of the methylation-specific probes showed a strong hypermethylation in the two patients and a slight hypomethylation of about 36% in the mother, who therefore carries the deletion on the paternal allele (Data for patient 3 not shown). (b) and (c) Sanger sequencing results for the deletion breakpoints. For patient 1 a 165 kb deletion comprising a 6 bp insertion between the proximal and distal breakpoints was observed (chr14:g.101285913_101451066del, hg19). For patients 2 and 3 as well as their mother a 5.8 kb deletion was present. The breakpoint junctions are uncertain at the triple C position (underlined; chr14:g.101291322_101297145del, hg19).
Initial genetic testing in patient 2 was performed by MS-PCR at the MEG3-DMR. Only the methylated paternal allele was present, microsatellite analysis showed biparental inheritance of chromosomes 14. A qPCR at the promoter region of MEG3 and a routine diagnostic genome-wide human SNP array analysis (resolution 100 kb) revealed no evidence for a deletion. MS-MLPA analysis for the 14q32 region was not available at that time. Thus it was concluded that the patient has a upd(14)pat syndrome probably owing to a primary epimutation. Five years later a sister (patient 3) was born with similar clinical features of a upd(14)pat syndrome. The clinical diagnosis could be confirmed by methylation analysis using a MS-SNuPE assay for the MEG3-DMR. A small deletion which was not detected before by genome-wide array analysis and qPCR could now be detected by MS-MLPA (Figure 3a) in both siblings: a 50% reduced dosage for three MLPA probes in the MEG3 promoter region was observed in the girl and her brother. A retrospective data analysis of the SNP array performed in patient 2 revealed that the deleted region has a low coverage of probes (2 probes) and therefore the deletion was not detectable by standard bioinformatic parameters. The same deletion was found in lymphocytes of the phenotypically unremarkable mother in a mosaic state with a signal reduction of ∼30% for the three probes. Long-range PCR was used to identify and further characterize the deletion breakpoints on the DNA sequence level. The deletion was found to be 5.8 kb in size encompassing the MEG3-DMR and the first three exons of the MEG3 gene (exon numbering based on NR_002766.2; hg19: chr14:g.101291322_101297145del; the patient has been submitted to LOVD (ID 00011680; http://databases.lovd.nl/) see Figures 1 and 3c).
Both, the 165 kb and the 5.8 kb deletion affect the MEG3-DMR but not the IG-DMR. Kagami et al19 reported a similar deletion of 4.3 kb including the MEG3-DMR, exon 1, exon 2 and part of exon 3, which does not affect methylation at the IG-DMR (Figure 3). In contrast, an 8.6 kb deletion including the IG-DMR but not the MEG3-DMR reported by the same group in another patient was associated with hypermethylation of the MEG3-DMR in blood DNA.
Since all applied tests investigated the methylation status of the MEG3-DMR but not the IG-DMR, we applied deep bisulfite sequencing of the MEG3-DMR and the IG-DMR using the Roche 454/GS junior system in our three patients to obtain high resolution quantitative DNA methylation patterns for the both regions. We also included the patient with the similar MEG3 deletion described by Kagami et al19 (patient with the 4.3 kb deletion), the parents of our cases 2 and 3, four normal controls, a patient with upd(14)pat and a patient with upd(14)mat in the analysis. Analysis of the sequence reads was performed using the open source amplikyzer software. As expected for the MEG3-DMR, all patients with a MEG3 deletion and the patient with upd(14)pat showed relative hypermethylation of 92 to 98% owing to the absence of the unmethylated maternal allele and the patient with upd(14)mat showed relative hypomethylation of 8.2% owing to the absence of the methylated paternal allele (Figure 4a). The mother of patients 2 and 3, who has the deletion in a mosaic state, was found to have an average methylation level of 36.8%. Since a normally methylated cell with two intact alleles for an imprinted gene has a methylated and an unmethylated allele and therefore a methylation level of ∼50%, the mother has ∼74% normally methylated cells and 26% cells carrying the deletion on the methylated paternal MEG3 copy. This is in agreement with the MS-MLPA results obtained in the gene dosage analysis.
Comparative methylation heatmaps for (a) the MEG3-DMR and (b) the IG-DMR. Each square represents a CpG dinucleotide with its average methylation level, each line a specific sample. The average methylation over the analyzed region is given in percentage under the sample ID together with the number of analyzed reads in brackets. The methylation was analyzed over 11 CpGs at the MEG3-DMR and over 15 CpGs at the IG-DMR. P indicates patient; * indicates patient with the 4.3 kb deletion described by Kagami et al.19
For the IG-DMR we investigated 15 of 19 CpG dinucleotides of the CG6 region, which has been analyzed by Kagami et al.19 via bisulfite cloning and sequencing. Between 1200 and 3200 sequence reads each for the 12 analyzed individual samples were obtained. Interestingly, in the normal controls only three of the 15 CpGs (CpG6-8, see Figure 4b) investigated showed a methylation level of about 50% which is the expected value for an allele-specifically methylated region at an imprinted locus. All other 12 CpGs studied showed an increased methylation level with an average methylation between 61 and 74%. Furthermore, the upd(14)mat sample showed 6–13% methylation for CpG6-8 and an overall methylation average of 32%, indicating that the maternal allele is not completely unmethylated. This is in agreement with the methylation pattern obtained by bisulfite Sanger sequencing in an upd(14)mat DNA sample shown by Kagami et al.19
For most of the CpGs studied, the deletion patients did not differ from the normal controls. But interestingly, all four patients including the patient with the 4.3 kb deletion published by Kagami et al.19 showed a nearly identical pattern of hypermethylation at certain CpGs, especially at CpG 3, 11 and 14, compared with all normal controls (Figure 4b). The mother of our cases 2 and 3, however, showed a normal methylation pattern at all CpGs.
Discussion
Based on the findings in two single patients with small deletions of maternal origin affecting only one of the imprinting regulatory elements in 14q32, the IG-DMR or the MEG3-DMR, it has been proposed that the IG-DMR acts as an upstream regulator for the methylation pattern of the MEG3-DMR. According to this hypothesis, a maternal deletion of the IG-DMR alone leads to a hypermethylation of the MEG3-DMR, whereas a maternal deletion of the MEG3-DMR alone does not change the methylation of the IG-DMR.19
We have now identified two additional patients with a deletion of the MEG3-DMR alone, one single patient with a maternal de novo microdeletion and two siblings with a microdeletion inherited from their mother who is a somatic mosaic for this deletion. By deep bisulfite sequencing we could confirm that, in general, these MEG3 deletions do not influence the methylation pattern of the IG-DMR, which strengthens the hypothesis of a hierarchical structure and distinct functional properties of the two DMRs.19 However, nearly identical changes in methylation levels were found at certain CpGs at the IG-DMR in our three patients plus the patient with the 4.3 kb deletion described by Kagami et al19 but not in any of the normal controls, suggesting that the MEG3 deletion on the maternal chromosome slightly influences the methylation at the IG-DMR. Interestingly we found that only three of the CpGs studied (CpGs 6–8), showed a methylation level of about 50% as expected for an allele-specifically methylated region at an imprinted locus. All other CpGs showed methylation >50%. The most likely explanation for this observation is that the maternal allele is not unmethylated in all cells. This is supported by the results obtained by deep bisulfite sequencing for the upd(14)mat sample which showed an average methylation level of 32% and also by the methylation pattern reported by Kagami et al19 in a upd(14)mat sample using bisulfite Sanger sequencing.
In the mother of cases 2 and 3 the deletion affects the paternally methylated MEG3-DMR and the silenced MEG3 gene copy, but not the methylation pattern at the IG-DMR. In contrast to the mother of the patient described by Kagami et al19 (patient with the 8.6 kb deletion), who has a deletion affecting the IG-DMR on the paternal chromosome and who has clinical features of upd(14)mat syndrome, the mother of our cases 2 and 3 has no phenotypic features of upd(14)mat. This suggests that the MEG-DMR on the paternal copy does not contain a cis-acting regulatory element for DLK1 expression at least within the deleted region as assumed for the IG-DMR19 or that the degree of cells carrying the deletion owing to the mosaic situation in the mother is too low to lead to a phenotypic effect. There is no comparable mouse model with a similar deletion. Only one mouse model with a targeted deletion upstream of the Meg3 gene has been reported so far.23 It could be shown that after paternal but also after maternal transmission mice carrying this deletion are indistinguishable from wild-type animals. They showed no phenotype and no change in the expression and imprinting of Dlk1 or Meg3. However, this deletion spans a 2.8 kb upstream region and leaves the Meg3 gene, Meg3 promoter and all but 100 bp of the Meg3-DMR intact and therefore it is not comparable with the deletion in our family.
Molecular genetics and clinical evaluations of patients and families revealed that almost all of the deletions affecting the IG-DMR and/or MEG3-DMR are associated with a high-recurrence risk for a upd(14)pat or a upd(14)mat syndrome, depending on the parental origin of the deletion (for review see Ogata et al8). Based on the absence of any phenotypic feature of upd(14)mat in the mother of our cases 2 and 3, it seems that a deletion restricted to the MEG3-DMR and affecting the paternal allele can be compatible with a normal phenotype. However, further patients are needed to elucidate the pathogenicity of such a deletion after paternal transmission as this is only a single case and since the mother is a somatic mosaic for the deletion.
As the family of cases 2 and 3 illustrates, it is important to have a reliable molecular genetic testing system to identify especially small deletions because of the high-recurrence risk. After the birth of the first child in our family (patient 2) we could not find evidence for a deletion (i) the sequence studied by qPCR maps 0.6 kb centromeric to the proximal deletion breakpoint and therefore failed to detect the deletion; (ii) owing to the small size of the deletion, the low coverage of probes in the deletion region (two SNPs) and the application of standard bioinformatic parameters used, the deletion escaped the detection by Genome-Wide Human SNP 6.0-Array analysis. No other testing system for this region was available at that time. Only after the development of a MS-MLPA containing several probes for the MEG3 locus, the deletion could be identified. To avoid false negative-molecular results in the diagnosis of patients with a clinical diagnosis of upd(14)mat or upd(14)pat syndrome it is necessary to improve and adopt the diagnostic strategies and methods to the current knowledge of molecular defects leading to the disease.
In summary, we could confirm the hypothesis of a hierarchical interaction and distinct functional properties of the IG-DMR and the MEG3-DMR in imprinting control of the 14q32 region.19 With the deep molecular analysis in patients with a maternal deletion of only the MEG3-DMR the functional predominance of the IG-DMR could be proven. The family with patients 2 and 3 demonstrated the need for a detailed characterization of molecular alterations in 14q32 for accurate genetic counseling.
References
Wylie AA, Murphy SK, Orton TC, Jirtle RL : Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation. Genome Res 2000; 10: 1711–1718.
Charlier C, Segers K, Wagenaar D et al: Human-ovine comparative sequencing of a 250-kb imprinted domain encompassing the callipyge (clpg) locus and identification of six imprinted transcripts: DLK1, DAT, GTL2, PEG11, antiPEG11, and MEG8. Genome Res 2001; 11: 850–862.
Cavaille J, Seitz H, Paulsen M, Ferguson-Smith AC, Bachellerie JP : Identification of tandemly-repeated C/D snoRNA genes at the imprinted human 14q32 domain reminiscent of those at the Prader-Willi/Angelman syndrome region. Hum Mol Genet 2002; 11: 1527–1538.
Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaille J : A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res 2004; 14: 1741–1748.
Paulsen M, Takada S, Youngson NA et al: Comparative sequence analysis of the imprinted Dlk1-Gtl2 locus in three mammalian species reveals highly conserved genomic elements and refines comparison with the Igf2-H19 region. Genome Res 2001; 11: 2085–2094.
Lin SP, Youngson N, Takada S et al: Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet 2003; 35: 97–102.
Kagami M, Sekita Y, Nishimura G et al: Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet 2008; 40: 237–242.
Ogata T, Kagami M, Ferguson-Smith AC : Molecular mechanisms regulating phenotypic outcome in paternal and maternal uniparental disomy for chromosome 14. Epigenetics 2008; 3: 181–187.
Cotter PD, Kaffe S, McCurdy LD, Jhaveri M, Willner JP, Hirschhorn K : Paternal uniparental disomy for chromosome 14: a case report and review. Am J Med Genet 1997; 70: 74–79.
Kagami M, Nishimura G, Okuyama T et al: Segmental and full paternal isodisomy for chromosome 14 in three patients: narrowing the critical region and implication for the clinical features. Am J Med Genet A 2005; 138A: 127–132.
Temple IK, Cockwell A, Hassold T, Pettay D, Jacobs P : Maternal uniparental disomy for chromosome 14. J Med Genet 1991; 28: 511–514.
Pentao L, Lewis RA, Ledbetter DH, Patel PI, Lupski JR : Maternal uniparental isodisomy of chromosome 14: association with autosomal recessive rod monochromacy. Am J Hum Genet 1992; 50: 690–699.
Antonarakis SE, Blouin JL, Maher J, Avramopoulos D, Thomas G, Talbot CC Jr : Maternal uniparental disomy for human chromosome 14, due to loss of a chromosome 14 from somatic cells with t(13;14) trisomy 14. Am J Hum Genet 1993; 52: 1145–1152.
Mitter D, Buiting K, von Eggeling F et al: Is there a higher incidence of maternal uniparental disomy 14 [upd(14)mat]? Detection of 10 new patients by methylation-specific PCR. Am J Med Genet A 2006; 140: 2039–2049.
Temple IK, Shrubb V, Lever M, Bullman H, Mackay DJ : Isolated imprinting mutation of the DLK1/GTL2 locus associated with a clinical presentation of maternal uniparental disomy of chromosome 14. J Med Genet 2007; 44: 637–640.
Buiting K, Kanber D, Martin-Subero JI et al: Clinical features of maternal uniparental disomy 14 in patients with an epimutation and a deletion of the imprinted DLK1/GTL2 gene cluster. Hum Mutat 2008; 29: 1141–1146.
Hosoki K, Ogata T, Kagami M, Tanaka T, Saitoh S : Epimutation (hypomethylation) affecting the chromosome 14q32.2 imprinted region in a girl with upd(14)mat-like phenotype. Eur J Hum Genet 2008; 16: 1019–1023.
Zechner U, Kohlschmidt N, Rittner G et al: Epimutation at human chromosome 14q32.2 in a boy with a upd(14)mat-like clinical phenotype. Clin Genet 2009; 75: 251–258.
Kagami M, O'Sullivan MJ, Green AJ et al: The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet 2010; 6: e1000992.
Dietz LG, Wylie AA, Rauen KA, Murphy SK, Jirtle RL, Cotter PD : Exclusion of maternal uniparental disomy of chromosome 14 in patients referred for Prader-Willi syndrome using a multiplex methylation polymerase chain reaction assay. J Med Genet 2003; 40: e46.
Begemann M, Leisten I, Soellner L, Zerres K, Eggermann T, Spengler S : Use of multilocus methylation-specific single nucleotide primer extension (MS-SNuPE) technology in diagnostic testing for human imprinted loci. Epigenetics 2012; 7: 473–481.
Beygo J, Citro V, Sparago A et al: The molecular function and clinical phenotype of partial deletions of the IGF2/H19 imprinting control region depends on the spatial arrangement of the remaining CTCF-binding sites. Hum Mol Genet 2013; 22: 544–557.
Steshina E, Carr M, Glick E, Yevtodiyenko A, Appelbe O, Schmidt J : Loss of imprinting at the Dlk1-Gtl2 locus caused by insertional mutagenesis in the Gtl2 5' region. BMC Genetics 2006; 7: 44.
Acknowledgements
We thank the families for their cooperation. Furthermore we wish to thank Melanie Heitmann and Christina Lich for expert technical assistance and Bernhard Horsthemke for helpful discussion and critical reading of the manuscript. Part of this study was supported by the Bundesministerium für Bildung und Forschung (Network Imprinting diseases, 01GM1114). ME, MB, GGK and TE are members of the European Network of Congenital Imprinting Disorders (EUCID.net), supported by COST (BM1208).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Beygo, J., Elbracht, M., de Groot, K. et al. Novel deletions affecting the MEG3-DMR provide further evidence for a hierarchical regulation of imprinting in 14q32. Eur J Hum Genet 23, 180–188 (2015). https://doi.org/10.1038/ejhg.2014.72
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2014.72
This article is cited by
-
Kagami Ogata syndrome: a small deletion refines critical region for imprinting
npj Genomic Medicine (2024)
-
Imprinting disorders
Nature Reviews Disease Primers (2023)
-
First step towards a consensus strategy for multi-locus diagnostic testing of imprinting disorders
Clinical Epigenetics (2022)
-
Novel parent-of-origin-specific differentially methylated loci on chromosome 16
Clinical Epigenetics (2019)
-
Maternally inherited 133kb deletion of 14q32 causing Kagami–Ogata syndrome
Journal of Human Genetics (2018)