Abstract
The present-day population structure of La Gomera is outstanding in its high aboriginal heritage, the greatest in the Canary Islands. This was earlier confirmed by both mitochondrial DNA and autosomal analyses, although genetic drift due to the fifteenth century European colonization could not be excluded as the main factor responsible. The present mtDNA study of aboriginal remains and extant samples from the six municipal districts of the island indeed demonstrates that the pre-Hispanic colonization of La Gomera by North African people involved a strong founder event, shown by the high frequency of the indigenous Canarian U6b1a lineage in the aboriginal samples (65%). This value is even greater than that observed in the extant population (44%), which in turn is the highest of all the seven Canary Islands. In contrast to previous results obtained for the aboriginal populations of Tenerife and La Palma, haplogroups related to secondary waves of migration were not detected in La Gomera aborigines, indicating that isolation also had an important role in shaping the current population. The rugged relief of La Gomera divided into several distinct valleys probably promoted subsequent aboriginal intra-insular differentiation that has continued after the European colonization, as seen in the present-day population structure observed on the island.
Similar content being viewed by others
Introduction
Islands can be considered as natural laboratories for studying the effect of founder events, drift or geographic isolation on the genetic composition of incoming colonizers. La Gomera is roughly circular, ~22 km in diameter and 370 sq.km in area. In spite of its small size, its central summit, Los Altos de Garajonay, reaches 1487 m above sea level. In addition, most of the island surface is >800 m above sea level with vertical cliffs around the coast, interrupted only by several narrow valleys that radiate from the centre to the sea. This rugged topography has hindered human travel within the island, as witnessed by the persistence till today of a whistled language used for communication over long distances.1 Sea travel was also difficult in the past. In fact, in contrast to archaeological evidence found on other islands of the archipelago, secondary intra-insular contacts of the pre-Hispanic aboriginals with La Gomera have not been demonstrated.2
Genetic evidence has pointed to a North African origin for the indigenous population, as well as a relatively high survival of the aboriginal gene pool in the modern mixed population. The strongest indigenous heritage has been observed in La Gomera, despite the high mortality rate of its aboriginal inhabitants during the European conquest in the fifteenth century.3 In previous AB0 system studies, La Gomera shows the highest 0 allele frequency (78%) of all the archipelago both at serological4, 5 and molecular6, 7 levels, resembling those found in aboriginal remains7, 8 and in North African Berber populations.6, 9, 10 Moreover, specific haplotypes of the CD4/Alu system also relate La Gomera to North African populations.11, 12, 13 These autosomal putative connections have been recently confirmed by a study using a wide number of informative ancestry markers, which estimated a remarkably higher North African ancestry for La Gomera (43%) compared with other islands.14
Without doubt, mtDNA has provided the most significant proof of the high aboriginal maternal heritage in the present day population of La Gomera. In a mtDNA study on the Macaronesian archipelagos, La Gomera behaved as an outlier compared with the rather homogenous Canarian group.15 This was mainly due to the fact that La Gomera has the highest frequency of the U6b1a mtDNA haplogroup (37%) compared with the other Canary Islands (~10%). This haplogroup is considered an aboriginal founder lineage,16 which has been fully confirmed by subsequent studies on aboriginal remains from Tenerife17 and La Palma.18 However, a Y-chromosome analysis of the current population was less conclusive, and genetic drift and/or sex-biased aboriginal gene flow to the postconquest population were proposed as the best explanation for these results.19, 20
The objective of this study was to directly assess the temporal evolution of the maternal aboriginal pool of La Gomera using mtDNA analysis on geographically ascribed samples from pre-Hispanic aboriginal remains and present-day samples collected from the six municipalities of the island.
Materials and methods
Sample collection
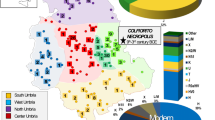
Prehistoric samples were obtained from 10 different archaeological sites on La Gomera (Table 1; Figure 1; Supplementary Table S1). A total of 97 different individual samples (101 teeth) were collected avoiding direct handling. Teeth on the same mandible were available in four cases only and were replicated independently at the Las Palmas laboratory following the same protocols as for La Laguna. Samples from Vallehermoso were 14C dated from 1600 to 1800 years before present.21 Stratigraphic level and burial characteristics confirmed the aboriginal nature of the rest of the samples.
Geographical distribution of municipalities and archaeological sites sampled in this study. Population codes are as in Table 1.
A total of 326 maternally unrelated modern samples were also collected from the six municipalities of La Gomera (Figure 1): Alajeró (ALA), Agulo (AGU), Vallehermoso (VH), San Sebastián (SS), Hermigua (HER), and Valle Gran Rey (VGR). Informed consent and the birthplaces of all their known ancestors were anonymously facilitated. Municipality assignation was established only when the maternal ancestry of the donors was confirmed until the third generation; in the remaining cases (41), they were ascribed only to the total sample of La Gomera. Previous samples of the current population of La Gomera used by Rando et al16 and Santos et al15 were retrieved, giving a total sample size of 398 (Supplementary Table S2). This study was approved by the Research Ethics Committee of the University of La Laguna.
DNA extraction
Ancient DNA (aDNA) was extracted as previously detailed.18 Briefly, after external decontamination using a 15% HCl solution and UV, teeth samples were extracted by means of a GuSCN-silica based protocol. In addition, re-extractions from the first pellet were carried out by dissolving it with 1.0 ml of GuSCN and repeating the extraction protocol. Modern DNA extractions from buccal swabs followed a protocol based on the use of proteinase K, dithiothreitol and sodium dodecyl sulphate.22
Real-time PCR quantification
Sample mtDNA quantification was performed on a 7500 Real Time PCR system (Applied Biosystem, Foster City, CA, USA) using a human-specific mtDNA fluorescent probe following the protocol published by Almeida et al.23
Amplification
To sequence the mtDNA hypervariable region I (HVRI) on ancient samples, we applied a previously published protocol.17 Briefly, we amplified a 400-bp HVRI region (from 16 000 to 16 400) using seven overlapping fragments, with sizes ranging from 82 to 124 bp. To improve sequencing, universal M13 primers (Promega, Sunnyvale, CA, USA) were added to the 5′ end of either forward or reverse primers and were used as a sequencing primer.18 In addition, ancient samples were analysed for several SNPs by RFLP analysis, to further characterize haplogroups H, JT, L and UK and subhaplogroups H1 and H3, using primers and conditions described elsewhere.17, 18 When all the aDNA analyses were finished, DNA was extracted, and the mtDNA HVRI region was analysed in present-day samples at a physically distant laboratory following a previously published protocol.24
Cloning
Ancient DNA amplification products were cloned into pGEM-T vectors (Promega), following the manufacturer’s conditions. Several clones were sequenced for each fragment until an unambiguous consensus sequence was obtained.17
Sequencing
aDNA PCR fragments were directly sequenced for both strands using the universal M13 forward primer (Promega) and the fragment-specific primer, while the M13 reverse primer was used for the clones as detailed previously.18 Modern samples were sequenced as in González et al.24
Criteria of authenticity
aDNA results were authenticated through the application of the following criteria: (a) lab-coats, face-shields, hats and sterile gloves were used at all times by both archaeological and genetic researchers during sample manipulation; (b) DNA extractions and amplification reactions were carried out in laminar flow cabinets at two independent laboratories exclusively dedicated to the study of aDNA, constantly irradiated with UV lamps when not in use; (c) the mtDNA control region was amplified using seven small overlapping fragments, increasing the probability of amplifying authentic aDNA; (d) independent extraction, amplification and sequencing of samples from the same individual were replicated in a second laboratory by a different researcher; (e) the absolute mtDNA copy number in aDNA extracts was assessed by real-time PCR; (f) a PCR reaction with the pair of primers that amplify the whole HVRI was tested for all the samples, and negative results were obtained; (g) one blank extraction per sample was processed in parallel, as well as three negative controls per PCR reaction; (h) eternal and internal DNA extractions from the same sample18 were carried out several times, to respectively assess the effectiveness of our decontamination process and contrast the different sources of the exogenous and endogenous DNA when both gave amplification products; (i) in 44.2% of the samples, at least two HVRI fragments were confirmed from a second re-extraction from the same individual; (i) in cases of heteroplasmy, PCR products were cloned to identify damage patterns or contaminant sequences; (k) haplogroup classification of haplotypes was confirmed by typing diagnostic SNPs; (l) the haplotypes obtained for ancient samples were compared with a panel of haplotypes from all the researchers involved in the study; (m) our overlapping amplicon approach allows the detection of any amplification of mtDNA sequences integrated into the nuclear genome; and (n) although there were no associated faunal remains to be analysed, we had previously extracted mtDNA from ancient lizard remains. These samples were obtained from caves with similar environmental conditions as those for the aboriginal burials but with older ages than those estimated for La Gomera samples.25
Statistical analysis
DNA sequences were analysed with the FinchTV program (Perkin Elmer, Buckinghamshire, UK) and confirmed by manually inspecting electropherograms. Special care was taken to detect heteroplasmic positions in aDNA sequences. Alignment was performed with the BioEdit software.26 Sequences were sorted into haplogroups following the updated nomenclature (mtDNA tree build 16; 16-2-2014) proposed by van Oven and Kayser.27 Aboriginal haplotypes were compared with those of the personnel involved in the excavation and manipulation of the aboriginal samples (Supplementary Table S3). Published samples used for comparisons are detailed in Supplementary Table S2. Gene diversity was calculated according to Nei.28 AMOVA analysis based on haplogroup frequencies was calculated using the Arlequin 3.5 software.29 Distances between populations were estimated using the Dest value,30 calculated by means of the R software31 as in Pennings et al.32 Principal Component Analysis (PCA) and Multidimensional Scaling (MDS) were performed using the R software. Admixture estimates were calculated with the WLSAdmix program33 (kindly provided by Dr Jeffrey Long).
Results
Authentication of ancient DNA samples
Ancient DNA re-extractions were successful in 92.3% of the samples compared with the first extraction. DNA yields of re-extractions were on average 66.6% of the previous extraction but with values ranging from 94.6 to 37.1%. Although we assayed various alternating extraction methods such as phenol/chloroform followed by the GuSCN-silica protocol and vice versa, the efficiency was similar to the currently used two consecutive GuSCN-silica extractions. This additional input of DNA was used to repeat genetic analysis of the same individual and to complete haplotypes of several samples when the first extract was finished. We were able to retrieve the sequence of the haplotypes from the anthropologist in charge of the physical measurements on the external tooth extraction in five instances but never in the endogenous extracts. Real-time PCR quantification allowed us to set the mean number of molecules used as initial PCR template at 2500–3000 copies. When the number of molecules obtained in the extractions was <250–100 copies/μl and further DNA concentration was not possible due to unsolvable inhibition problems, the samples were discarded for the genetic analysis. Cloning was performed on 17 fragments in which heteroplasmy was detected, and in all cases, sequences with mutations included in the haplotypes of both researchers engaged in the molecular analysis were retrieved as well as some mutations attributed to miscoding lesions: 16014, 16219, 16232, and 16355 (Supplementary Table S4). Both-strand sequencing allowed the identification of the following phantom mutations: 16067, 16270, 16297, 16302, 16304A, 16316, and 16325. The 4 samples and 10 re-extractions replicated in Las Palmas gave identical results to those in La Laguna. In summary, 37 samples had to be discarded because of poor DNA yield (38.1%), unsolvable inhibition (3; 3.1%) or extraction contamination (5; 5.1%), while 52 informative mtDNA sequences were obtained from a total of 97 samples—an overall efficiency of 53.6%.
Aborigine mtDNA structure
At the time of the European conquest, La Gomera was subdivided into four political cantons named Hipalán, Mulagua, Orone and Agana, with some geographical correspondence to the current municipalities of San Sebastián, Hermigua-Agulo, Alajeró-Valle Gran Rey and Vallehermoso, respectively (Figure 1). Although during the fifteenth century an East–West boundary was politically recognized, it is known that previously a North–South boundary existed.34 The 52 aboriginal sequences analysed gave a total of 10 different haplotypes assorted into 8 haplogroups (Table 1), giving a whole gene diversity of only 55.88±7.75%. Subdivision of the samples into northern and southern areas showed that the autochthonous U6b1a haplogroup was overwhelmingly dominant on the island (Table 2). However, there were significant differences between areas due to the exclusive presence of haplogroup J lineages in the South (P=0.039) and haplogroup L lineages in the North (P=0.029). These results point to geographical genetic differentiation on La Gomera before the European conquest, although we cannot exclude that these differences could be related to small sample sizes or temporal differences during pre-Hispanic times.
Extant mtDNA structure
Although comparatively low, the present-day gene diversity on La Gomera (78.21±1.97%) is significantly higher than that of the aborigines (Table 2). Moreover, the presence of European, sub-Saharan African and Amerindian new lineages testify to the impact of the European colonization despite the autochthonous U6b1a haplogroup (44%) remaining dominant. (Table 2; Supplementary Table S5). AMOVA analysis showed that about 2% of the variance is due to significant (P=0.002) differences among municipalities. In particular, Alajeró was characterized by the highest frequencies for H/HV/R (P<0.0001), T2c1 (P=0.0006) and U6c1 (P=0.0026) haplogroups and Agulo by those of L3e (P=0.001) and U6a (P=0.0044). T2b was particularly abundant (P<0.0001) in Hermigua, haplogroup K1b1a was detected only in San Sebastián (P=0.0456), while L3d (P=0.0033), T1 (P=0.0047) and W (P=0.0012) were more frequent in Valle Gran Rey. Furthermore, Vallehermoso and Agulo harboured the highest frequencies of haplogroup J (P=0.0027) and Alajeró and Valle Gran Rey those of haplogroup I (P=0.022). It should be noted that the Canary indigenous founder lineages16, 17, 18 T2c1, U6a and U6c1 were found in the extant population of La Gomera but are absent in the aboriginal sample. This could be the result of Canary Islanders moving to the island during and after the conquest; however, historical records speak of greater movement of aborigines from La Gomera to Tenerife.35 Excluding L lineages already present in the aboriginal sample, the total sub-Saharan African contribution to La Gomera’s population, most probably due to the postconquest slave trade, was only moderate (6.5%).
Temporal discontinuity
Although a high aboriginal input was observed, it has evolved under the influence of the conquest and the strong migratory European input. The Canarian U6b1a haplogroup has dropped from 61–71% in pre-Hispanic times to 40–50% in the present-day La Gomera population (P=0.0047). Haplogroup X is another example of decline through time as it was relatively abundant in the indigenous population (11–4%) but is now barely present (0.25%) on the island (P=0.0007). Some haplogroups not detected in some areas before the conquest are now especially abundant. For example, haplogroup J now has a frequency of 12.5% in the North of the island, but it was not detected in the northern aboriginal sample (P=0.033). On the contrary, haplogroup L3d was significantly more abundant in the North before the conquest than it is in the current population (P=0.0125). Pair-wise distances comparing the aboriginal and extant samples show that the southern aboriginal sample was strongly differentiated from the northern and from all present-day samples. In contrast, the northern pre-Hispanic population was comparatively more similar to the extant Gomeran samples, paradoxically showing its closest affinity with the southern municipality of Alajeró (Supplementary Table S4).
Discussion
Aboriginal inter-insular differences
Gene diversity (55.9±7.8%) in aborigines from La Gomera (known as Gomeros) was significantly lower than those previously found for the indigenous Canary Island people in general. Specifically, aborigines from Tenerife (known as Guanches) and from La Palma (known as Benahoaritas) showed diversity values within the range of current insular populations (77.5±6.1%17 and 67.6±9.3%,18 respectively). Compared with the other islands, these data point to a stronger founder effect during the aboriginal colonization of La Gomera and/or a greater subsequent isolation. The most striking differences observed between islands are the high frequency of the autochthonous haplogroup U6b1a in Gomeros (around 65%) compared with Guanches (11%) and Benahoaritas (7%) and the low frequency of haplogroup H (around 8%) in La Gomera compared with the other two aboriginal populations (44 and 57%, respectively). The lack of the founder haplogroup T2c1 in Gomeros and its frequency in Guanches (14%) and Benahoaritas (10%) is also outstanding. Gomeros also lack L2 lineages, which are present in both Guanches (8%) and Benahoaritas (3%). Particular similarities of aborigines from La Gomera with those of La Palma are the exclusive sharing of haplogroups X and L1b (Table 2), and with Tenerife the sharing of L3d haplotypes (not found in La Palma) (Table 2). However, at least some of these differences could be attributed to small sample sizes. In general, the sample of southern Gomeros showed comparatively fewer differences with Guanches and Benahoaritas than the northern Gomeros. Moreover, Guanches were more similar to the present-day Gomeran population than were Benahoaritas (Supplementary Table S6). This would be in agreement with archaeological findings relating La Gomera and Tenerife aboriginal cultures.
It has been estimated that the potential founder lineages such as U6b1a or H1-16260 were more ancient than the oldest archaeological site in the Canary Islands, dated at the first millennium BC.18, 36 To reconcile these discrepancies, it was suggested that more than one haplotype per founder haplogroup might have arrived in the Canary Islands with the aboriginal colonization.37 The presence in Gomeros of the autochthonous U6b1a-derived haplotypes 048 163 172 219 311 and 092 163 172 219 311, in addition to the basal 163 172 219 311 (Table 1), would support the idea that these derived lineages did not mutate in the Canary Islands but arrived there from North Africa with the aboriginal colonization. However, this hypothesis should be reviewed if older archaeological remains are discovered in the islands.
Singularity of the extant population of La Gomera
It has been previously stated that the extant population of La Gomera was genetically differentiated from other islands of the archipelago.15, 16 This difference was attributed to a better preservation of the maternal aboriginal substrate, although the effect of genetic drift could not then be ruled out. Now, we know that the comparatively high presence of the autochthonous haplogroup U6b1a in the current population of La Gomera has been inherited from the aboriginal people and was not caused by later genetic drift.
The subdivision of the total Gomeran population into municipalities allows refined comparisons with other islands. Tenerife is the population with the lowest genetic distance from La Gomera, the municipalities Agulo, Hermigua and Valle Gran Rey being the closest (Supplementary Table S4). This result is congruent with the geographical proximity of La Gomera to Tenerife (Figure 1). When all the islands are taken into account, the first component of PCA analysis (accounting for 17.2% of the variability) shows that the Gomeran municipalities and all the indigenous population are clustered to the left, whereas the other Canary Islands samples, including the eighteenth century population of Tenerife, are clustered to the right (Figure 2a). Congruently, San Sebastián, the main port and capital of La Gomera and therefore the most affected by external gene flow, is the closest to the right cluster. The main haplogroups responsible for the separation of the two clusters are the European U5b and H to the right and the Canarian autochthonous U6b1a to the left. It is clear that the aboriginal samples of Tenerife and La Palma are situated in the left cluster due to the strong indigenous component of the extant and prehistoric samples from La Gomera. The second component (12.8%) further separates San Sebastián and, to a lesser degree, Gran Canaria and Tenerife from the other populations, through their comparatively higher frequency of U5b2, L2a and K1b1a lineages. The MDS plot based on Dest distance values (Figure 2b) indicates that all the modern municipalities of La Gomera and the northern aboriginal samples are similar to each other and different from the cluster conformed by the rest of the archipelago, including the aboriginal samples from La Palma and Tenerife. In this case, the southern aboriginal sample from La Gomera behaves as an outlier of the Gomeran cluster, probably due to its higher frequency of U6b1a (~71%).
(a) PCA results based on haplogroup frequencies. (b) MDS plot based on Dest distance values. Population codes are as in Table 2. Black and gray icons correspond to ancient and present-day populations, respectively. Circles correspond to Gomeran populations, while squares indicate other Canary Islands samples. Dark and light gray ovals indicate Gomeran and non-Gomeran clusters.
After the first analysis of the aboriginal samples,17 admixture estimates of the current population became calculable using the aboriginal sample as one of the parental contributors (Iberians and West sub-Saharan Africans being the others), instead of inferring them from North African Berber populations as before.16, 38 Mean estimates pointed to the Iberian Peninsula as the main contributor to the Canarian maternal gene pool with 55%, followed by 42% of aboriginal influence and a minimal sub-Saharan African input of around 3%.17 Repeating calculations for the whole population of La Gomera using Gomeros as indigenous parental population, we obtained percentages of 55.4, 37.8 and 6.8% for the aboriginal, Iberian and sub-Saharan African components, respectively, in this case the aboriginal substrate being the largest. Taking into account the different municipalities, this indigenous contribution ranges from 62.4% in Alajeró to 48.1% in San Sebastián (Supplementary Table S7). The Sub-Saharan input due to postconquest slavery varies from 0.0% in Alajeró to 15.6% in Agulo.
The comparison of mtDNA and Y-chromosome data from the current Canary Islands population16, 38 led to the detection of a significantly higher contribution of aboriginal matrilineages compared with patrilineages. This result is usually explained by the asymmetrical migration of European colonizers, which principally consisted of males. The reduction in the male aboriginal gene pool can also be explained both by their higher mortality rate during the conquest and their lower socioeconomic status afterwards.20 The case of La Gomera in particular, with the highest (around 50%) aboriginal maternal contribution, (Santos et al,15 Rando et al16 and this study) and one of the lowest (around 6%) aboriginal paternal contributions19 may indeed reflect the particularly brutal way this island was finally conquered, producing the strongest sexual asymmetry in the Archipelago.3 Alternatively, strong genetic drift could also be the main cause of this striking sexual asymmetry.19 Unfortunately, the low nuclear DNA yield of the Gomero sample prevented a direct analysis of its Y-chromosome profile. However, taking into account the aboriginal contribution estimated from autosomal data (43%)14 and the indigenous maternal contribution (55.2%) obtained here, applying the formula: mAUTO=1/2 m mtDNA+1/2 m NRY39 provides a theoretical aboriginal Y-chromosome ancestry of about 31%. This value is at least twice that obtained using current North African Berbers as parental population.40 As higher male mortality and asymmetrical mating would especially occur at the beginning of the colonization, we conclude that male extermination could have been less drastic than suggested and that male gene drift also had an important role.
Conclusion
Ancient mtDNA results from La Gomera samples are congruent with the phylogeographic pattern observed in the current population and with archaeological evidence. The high diversities and close affinities found previously for the aboriginal samples from Tenerife17 and La Palma18 better fit an island model of aboriginal colonization, with several arrivals and/or frequent migration between islands.18 In contrast, the low diversity, strong intra-insular microdifferentiation and high inter-insular heterogeneity found in the aboriginal samples from La Gomera appears to reflect a single colonization process, with a strong founder event and subsequent isolation. This idea is strongly reinforced by the absence in the aboriginal Gomeran samples of the founder lineages T2c1 and U6c1, proposed markers of a secondary pre-Hispanic colonization elsewhere in the islands.15 The abrupt topography of the island must also have promoted isolated settlements of the Spanish colonizers in historical times that, after mixing with the local aborigines, kept relatively segregated until recent times. This would also contribute to the strong genetic structure found in La Gomera. In conclusion, this small island is not only particularly rich in endemic flora and fauna but is also where the pre-Hispanic aboriginal substrate is best preserved in the Canary archipelago.
References
Carreiras M, Lopez J, Rivero F, Corina D : Linguistic perception: neural processing of a whistled language. Nature 2005; 433: 31–32.
Navarro JF: Arqueología de las Islas Canarias. Espacio, Tiempo y forma Serie I, Prehistoria y Arqueología, Spain: Revista de la Facultad de Geografia e Historia de Madrid (UNED) 1997; 10: 447.
Suárez JJ, Rodriguez F, Quintero CL : Historia Popular de Canarias, Vol 2. Conquista y Colonización, Spain: Centro de la Cultura Popular Canaria, 1988.
Roberts DF, Evans M, Ikin EW, Mourant AE : Blood groups and the affinities of the Canary islanders. Man 1966; 1: 512–525.
Pinto F, Rando JC, Lopez M, Morilla JM, Larruga JM : Blood group polymorphisms in the Canary Islands. Gene Geogr 1996; 10: 171–179.
Fregel R, Maca-Meyer N, Cabrera VM, Gonzalez AM, Larruga JM : Description of a simple multiplex PCR-SSCP method for AB0 genotyping and its application to the peopling of the Canary Islands. Immunogenetics 2005; 57: 572–578.
Fregel R, Betancor E, Suarez NM et al: Temporal evolution of the ABO allele frequencies in the Canary Islands: the impact of the European colonization. Immunogenetics 2009; 61: 603–610.
Schwarzfischer F, Liebrich K : Serologische Untersuchungen an prähistorischen Bevölkerungen, insbesondere an altkanarischen Mumien. Homo 1963; 14: 129–133.
Gaud J : Contribution à l'étude ethnographique du Maroc. Répartition des groupes sanguins parmi les tribus des environs de Meknès. Bull Inst Hyg Maroc 1942; 2: 37–52.
Messerlin A, Lorho Y : Groupes sanguins et facteur Rhésus au Maroc. Bull Inst Hyg Maroc 1951; 11: 97–119.
Flores C, Maca-Meyer N, Gonzalez AM, Cabrera VM : Northwest African distribution of the CD4/Alu microsatellite haplotypes. Ann Hum Genet 2000; 64: 321–327.
Flores C, Maca-Meyer N, Perez JA, Cabrera VM : The peopling of the Canary Islands: a CD4/Alu microsatellite haplotype perspective. Hum Immunol 2001; 62: 949–953.
Maca-Meyer N, Villar J, Perez-Mendez L, Cabrera de Leon A, Flores C : A tale of aborigines, conquerors and slaves: Alu insertion polymorphisms and the peopling of Canary Islands. Ann Hum Genet 2004; 68: 600–605.
Pino-Yanes M, Corrales A, Basaldua S et al: North African influences and potential bias in case-control association studies in the Spanish population. PLoS One 2011; 6: e18389.
Santos C, Fregel R, Cabrera VM, Gonzalez AM, Larruga JM, Lima M : Mitochondrial DNA patterns in the Macaronesia islands: Variation within and among archipelagos. Am J Phys Anthropol 2010; 141: 610–619.
Rando JC, Cabrera VM, Larruga JM et al: Phylogeographic patterns of mtDNA reflecting the colonization of the Canary Islands. Ann Hum Genet 1999; 63: 413–428.
Maca-Meyer N, Arnay M, Rando JC et al: Ancient mtDNA analysis and the origin of the Guanches. Eur J Hum Genet 2004; 12: 155–162.
Fregel R, Pestano J, Arnay M, Cabrera VM, Larruga JM, Gonzalez AM : The maternal aborigine colonization of La Palma (Canary Islands). Eur J Hum Genet 2009; 17: 1314–1324.
Flores C, Maca-Meyer N, Perez JA, Gonzalez AM, Larruga JM, Cabrera VM : A predominant European ancestry of paternal lineages from Canary Islanders. Ann Hum Genet 2003; 67: 138–152.
Fregel R, Gomes V, Gusmao L et al: Demographic history of Canary Islands male gene-pool: replacement of native lineages by European. BMC Evol Biol 2009; 9: 181.
Arnay-de-la-Rosa M, Gamez-Mendoza A, Navarro-Mederos JF et al: Dietary patterns during the early prehispanic settlement in La Gomera (Canary Islands). J Archaeol Sci 2009; 36: 1972–1981.
Maniatis T, Fritsch EF, Sambrook J : Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York, USA: Cold Spring Harbor Laboratory, 1982.
Almeida M, Betancor E, Fregel R, Suárez NM, Pestano J : Efficient DNA extraction from hair shafts. Forensic Sci Int Genet Suppl Ser 2011; 3: e319–e320.
Gonzalez AM, Cabrera VM, Larruga JM et al: Mitochondrial DNA variation in Mauritania and Mali and their genetic relationship to other Western Africa populations. Ann Hum Genet 2006; 70: 631–657.
Maca-Meyer N, Carranza S, Rando JC, Arnold EN, Cabrera VM : Status and relationships of the extinct giant Canary Island lizard Gallotia goliath (Reptilia: Lacertidae), assessed using ancient mtDNA from its mummified remains. Biol J Linn Soc 2003; 80: 659–670.
Hall TA : BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 1999; 41: 95–98.
van Oven M, Kayser M : Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat 2009; 30: E386–E394.
Nei N : Molecular Evolutionary Genetics. New York, USA: Columbia University Press, 1987.
Excoffier L, Lischer HEL : Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 2010; 10: 564–567.
Jost L : G(ST) and its relatives do not measure differentiation. Mol Ecol 2008; 17: 4015–4026.
R Development Core Team R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, 2008. ISBN 3-900051-07-0, Available at http://www.R-project.org.
Pennings PS, Achenbach A, Foitzik S : Similar evolutionary potentials in an obligate ant parasite and its two host species. J Evol Biol 2011; 24: 871–886.
Long JC : The genetic structure of admixed populations. Genetics 1991; 127: 417–428.
Navarro JF : La Prehistoria de Canarias, Vol 5. La Gomera y Los Gomeros. Centro de la Cultura Popular Canaria: Santa Cruz de Tenerife, Spain, 1992.
Lobo-Cabrera M : Los Gomeros en el Poblamiento de Tenerife Vol. XLV. El Museo Canario: Las Palmas de Gran Canaria, Spain, 1984.
Pereira L, Silva NM, Franco-Duarte R et al: Population expansion in the North African late Pleistocene signalled by mitochondrial DNA haplogroup U6. BMC Evol Biol 2010; 10: 390.
Secher B, Fregel R, Larruga JM et al: The history of the North African mitochondrial DNA haplogroup U6 gene flow into the African, Eurasian and American continents. BMC Evol Biol 2014; 14: 109.
Pinto F, Gonzalez AM, Hernandez M, Larruga JM, Cabrera VM : Genetic relationship between the Canary Islanders and their African and Spanish ancestors inferred from mitochondrial DNA sequences. Ann Hum Genet 1996; 60: 321–330.
Lind JM, Hutcheson-Dilks HB, Williams SM et al: Elevated male European and female African contributions to the genomes of African American individuals. Hum Genet 2007; 120: 713–722.
Arnay-de-la-Rosa M, Gamez-Mendoza A, Navarro-Mederos J et al: Dietary patterns during the early prehispanic settlement in La Gomera (Canary Islands). J Archaeol Sci 2009; 36: 1972–1981.
Acknowledgements
This research would have not been possible without the enthusiastic collaboration of the anonymous donors from La Gomera and the support of the staff from Área de Salud de La Gomera (Centro Canario de Salud) and IES San Sebastián de La Gomera. This work was funded by the Spanish Ministerio de Ciencia e Innovación (CGL2010–16195 to AMG) and by the Universidad de La Laguna (Ayuda para el mantenimiento de grupos de investigación consolidados 2012, number 2012/1552 to AMG).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Fregel, R., Cabrera, V., Larruga, J. et al. Isolation and prominent aboriginal maternal legacy in the present-day population of La Gomera (Canary Islands). Eur J Hum Genet 23, 1236–1243 (2015). https://doi.org/10.1038/ejhg.2014.251
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2014.251
This article is cited by
-
Archaeological Research in the Canary Islands: Island Archaeology off Africa’s Atlantic Coast
Journal of Archaeological Research (2023)
-
The genomic history of the indigenous people of the Canary Islands
Nature Communications (2023)