Abstract
Genome-wide linkage analysis, followed by targeted deep sequencing, in a Danish multigeneration family with juvenile cataract revealed a region of chromosome 17 co-segregating with the disease trait. Affected individuals were heterozygous for two potentially protein-disrupting alleles in this region, in ACACA and UNC45B. As alterations of the UNC45B protein have been shown to affect eye development in model organisms, effort was focused on the heterozygous UNC45B missense mutation. UNC45B encodes a myosin-specific chaperone that, together with the general heat shock protein HSP90, is involved in myosin assembly. The mutation changes p.Arg805 to Trp in the UCS domain, an amino acid that is highly conserved from yeast to human. UNC45B is strongly expressed in the heart and skeletal muscle tissue, but here we show expression in human embryo eye and zebrafish lens. The zebrafish mutant steif, carrying an unc45b nonsense mutation, has smaller eyes than wild-type embryos and shows accumulation of nuclei in the lens. Injection of RNA encoding the human wild-type UNC45B protein into the steif homozygous embryo reduced the nuclei accumulation and injection of human mutant UNC45B cDNA in wild-type embryos resulted in development of a phenotype similar to the steif mutant. The p.Arg805Trp alteration in the mammalian UNC45B gene suggests that developmental cataract may be caused by a defect in non-muscle myosin assembly during maturation of the lens fiber cells.
Similar content being viewed by others
INTRODUCTION
Congenital/infantile cataract (CC) is a developmental anomaly characterized by opacities in the crystal lens of the eye and is a common cause of restricted vision and blindness in children. Environmental and intrinsic factors are involved, including metabolic and genetic causes for CC. Mendelian forms of CC comprise a broad spectrum of syndromic and nonsyndromic phenotypes characterized by a set of associated ocular and/or systemic abnormalities. More than 35 loci, including at least 25 known genes, have been associated with nonsyndromic cataract, the majority showing autosomal dominant inheritance with high penetrance (ADCC).1, 2 Mutations in crystallins, particularly CRYAA, CRYBB2, and CRYGD and the connexin genes GJA3 and GJA8 comprise the largest group of loci causing ADCC, but mutations are also found in the membrane proteins MIP, LIM2, TMEM114 and CHMP4B, in cytoskeleton proteins (BSFP1 and BSFP2) and in transcription factors (HSF4 and MAF).1, 2
In a recent study,3 we reported that these loci represented most of the causative mutations in 19 out of 28 unrelated Danish CC families. Among the unsolved families was one in which genome-wide linkage analysis revealed two candidate loci at 2q32.3-q33.3 and 17q11.2-q21.2, respectively.3 The family comprised three generations with nine affected members and the hereditary mode was consistent with autosomal dominant inheritance with complete penetrance (Figure 1a). Here we present the targeted resequencing of the linkage regions resulting in identification of a causative mutation in the myosin chaperone UNC45B. As zebrafish (Danio rerio) is a widely used model for human congenital disorders of the eye including cataracts,4 we show the zebrafish homolog is necessary for normal early lens development.
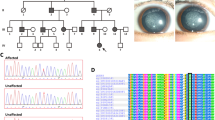
(a) The pedigree of family CC00116; filled symbols denote affected persons, circles denote females and squares denote males. (b) RT–PCR gene expression analyses of UNC45B, HSF4, ACACA, MYH9, MYH10, HSP90AA1 and HSP90AB1 in human embryo 43-day-old (1) and 54-day-old (2) eyes. The HSF4 transcript was represented by two transcript variants (NM_001040667.2 and NM_001538.3). (c) Schematic domain structure of the UNC-45 family of proteins depicts the tetratricopeptide repeat (TPR) Hsp90 interaction domain, the central UNC-45 domain and the UCS myosin interaction domain. The clustered mutations in the UCS domain are shown by arrows, and the positions of the lethal (p.Arg210* and p.Trp335*) and the temperature-sensitive (ts) mutations in the C. elegans30 homolog (p.Gly427Glu and p.Leu559Ser) are shown by an asterisk. The protein numbering refers to human UNC45B. (d) Structure of human UNC45B, central region and UCS domain based on the D. melanogaster 3D structure (PDB ID:3now).33 The five ARMR structures are shown in different colors with the conserved groove in the brown. The h-Glu768 and the h-Arg805 residues are shown by arrows. The α-helices structure is show in upper left corner. Phyre2 and FirstGlance34 were used for making the human model. (e) Alignment of the highly conserved subregion of the UCS domain with part of the α-helix structure for 18H–19H (modified from33). Altered amino-acid positions are colored red and the conserved inter-repeat interacting ion pair h-Glu768 and h-Arg805 residues (Glu766 and Arg803 in the D. melanogaster sequence) are denoted.
MATERIALS AND METHODS
The family CC00116 was recruited from The National Danish Register of Hereditary Eye Diseases at the National Eye Clinic, Kennedy Center (http://www.kennedy.dk/). The study adhered to the tenets of the Declaration of Helsinki and was approved by the Copenhagen Scientific Ethics Committee and after being informed, all subjects gave written consent to participate in the study. Retrospective clinical information was obtained from ophthalmologists in private practice and local ophthalmic hospital departments. Genome-wide linkage analysis was made using Affymetrix 10K SNP arrays (Affymetrix, Santa Clara, CA, USA), multipoint genetic linkage analysis and haplotyping was done using standard methods.5, 6 PCR, Sanger sequencing and diagnostic restriction enzyme analyses (New England Biolabs, Ipswich, MA, USA) were carried out according to standard protocols. The linkage regions (Supplementary Table S1) were captured in the affected individual I:1 (Figure 1a) using a NimbleGen custom-designed chip (Roche NimbleGen, Inc., Madison, WI, USA) and deep sequenced by paired-end tags using an Illumina Genome Analyzer IIx platform (Illumina, Inc., San Diego, CA, USA), and data were analyzed using standard protocols. The UNC45B cDNA clone NM_001033576 purchased from Origene (SC306792, Origene, Rockville, MD, USA) was used for site-directed mutagenesis. Total RNA was isolated from human embryo 43- and 54-day-old eyes and analyzed for gene expression by RT–PCR (Figure 1; Supplementary Information).
Zebrafish maintenance
Zebrafish were maintained and manipulated as described.7, 8, 9 The unc45b mutant allele steif (sb60) was generously provided by the Max-Planck-Institute für Entwicklungsbiologie (Tübingen, Germany).10 Zebrafish embryo genotypes were determined phenotypically or by using dCAPS analysis.11, 12
Immunohistochemistry and cryosection of Zebrafish embryonic eyes
Polyclonal antibodies were raised in guinea pigs against bacterially expressed recombinant zebrafish Unc45b protein. Cryosections of embryonic eyes were prepared and immunostained as described.13, 14 non-muscle myosin (NMM) antibody (NMMII;M8064, Sigma-Aldrich, Oakville, ON, Canada) was used at 1:100 dilution, whereas ZL-1 (Zebrafish International Resource Center, University of Oregon) or Unc45b was used at 1:250 dilution.
Ectopic Unc45b in zebrafish
Plasmids containing human wild-type and mutant UNC45B clones (described above) were linearized and UNC45B capped mRNA was synthesized using T7 polymerase (Invitrogen, Carlsbad, CA, USA) and the mMessage mMachine kit (Life Technologies Inc., Burlington, ON, Canada). Two hundred and fifty pg of mRNA was injected into embryos at the one-cell stage. Embryos were genotyped by dCAPS as described above. More than 20 embryos were injected in each trial.
RESULTS
Clinical data
The pedigree of the family with autosomal dominant CC is shown in Figure 1a. Clinical data (obtained retrospectively) were limited for this family, as most of the affected individuals had already been operated upon and were aphakic at the time of investigation. The age at first operation varied between 6 and 45 years of age (mean=19, median=14 (N=7)). In two individuals, retinal detachments occurred after operation for secondary cataract and one individual developed a severe secondary glaucoma. Individual III:4 (born in 1950) was diagnosed with cataract at the age of 18 and not operated before the age of 45. The phenotype was described as posterior subcapsular and central. During intraocular lens implantation, a central fibrous opacity in the lens capsule had to be left owing to a risk of rupture of the capsule. In the youngest generation, the two affected individuals were followed for years before lens opacities developed at ages 9 and 16, respectively.
NGS and characterization of the mutations
Genome-wide linkage analyses resulting in two candidate loci on chromosomes 2 and 17 has previously been published (Supplementary Figures S1A and S2).3 The linkage regions covered 24 Mbp and 254 annotated reference genes (Supplementary Table S1). Ten genes selected as candidates based on EST and cDNA expression data were all excluded by Sanger sequencing (Supplementary Figure S1B). Subsequently targeted deep sequencing of the two linkage regions in the index patient I:1 resulted in a total of 203 public (dbSNP138) and 5 private SNVs (Supplementary Table S1). One hundred and five public and four private SNVs were nonsynonymous variants. Sanger sequencing and segregation analyses excluded three of the four private nonsynonymous SNVs, leaving an ACACA mutation as a disease candidate. One hundred four of the 105 public nonsynonymous SNVs were excluded either by a MAF value >0.01 or by Sanger sequencing and segregations analysis (data not shown), leaving an UNC45B mutation as a putative disease-causing lesion. Five mutations in splice regions were excluded owing to high MAF values; gain or loss stop variants were not found in the exons (Supplementary Figure S1B and Supplementary Table S1). UNC45B and ACACA mutations segregated heterozygously with the disease trait and homozygous for the canonical sequence in unaffected members confirming autosomal dominant inheritance (Figure 1a).
The ACACA (MIM 200350) mutation was a G>A transversion (c.3398G>A) in exon 27 of transcript variant 1 causing a substitution of Arg1133 with His. The mutation affects the central region of the ACACA protein and was predicted to be damaging by the prediction servers PolyPhen2 (0.986), MutationTaster (0.79) and SIFT.
The UNC45B (MIN 611220) mutation was a C>T transition (c.2413C>T) in exon 19 of transcript variant 1 and replaces Arg805 with a tryptophan residue. The mutation is located in the in C-terminal UCS domain of the protein (Figure 1) and predicted damaging by PolyPhen2 (1.000), MutationTaster (1.000) and SIFT. The human UNC45B Arg805 residue is highly conserved in vertebrate as well as in invertebrate and fungal homologs (Supplementary Figures S3C and S3D).
Neither of the mutations was found by Sanger sequencing or restriction enzyme digest in 230 unrelated healthy Danes of both sexes. The ACACA substitution has been reported by the COSMIC project (Id: COSM175625) as a large intestine carcinoma somatic mutation, and the UNC45B mutation (rs370424081) by the NHLBI Exome Sequencing Project (ESP6500, November 2013) in the African American population (genotype frequencies TT=0/TC=1/CC=2202).
Gene expression data
To test whether the expression of either of the two candidate genes was consistent with a role in the lens, RNA from 43- and 54-day human embryonic eye tissues was used for qualitative PCR analyses of UNC45B and ACACA, the human HSP90 genes HSP90AA1 (MIN 140571) and HSP90AB1 (MIN 140572), the NMM heavy chain genes MYH9 (MIM 160775) and MYH10 (MIM 160776), and the cataract disease gene HSF4 (MIM 602438). Expression of UNC45B, ACACA, MYH9, MYH10, the two HSP90 genes and the HSF4 gene was detected at both developmental stages, which did not allow exclusion of either gene from consideration (Figure 1b).
Given that the ACACA gene is proposed to encode an enzyme with a housekeeping function, and is most likely to have an effect in highly lipogenic tissues, while the UNC45B locus has been shown in model systems to have both an eye phenotype when mutant as well as regulating the function of NMM in non-muscle tissues,10, 15 and heterozygous mutations in NMMs have been associated with premature cataract formation in mouse models,16 our attention focused on UNC45B as the more likely candidate gene. We cannot rule out the formal possibility that the CC phenotype in this family may result from contributions from multiple heterozygous loci.
Zebrafish studies
The Unc45b ortholog in zebrafish shows expression in the embryonic eye, both by antibody staining and by reporter gene (Figures 2a–c). Differentiation of lens fiber cells begins in the zebrafish at ∼24 h post fertilization (hpf) and involves processes including cellular reorganization and organelle degeneration that result in a transparent lens and functional visual system by 72 hpf.17, 18 Disassembly of actin stress fibers is believed to induce lens cell differentiation and lead to the formation of cortical fibers through the reorganization of actin filaments.18, 19 Zebrafish embryonic eyes were examined for lens fiber cell boundaries, differentiation, and nuclear retention. Expression of zebrafish Unc45b is seen both in the epithelial layer of the developing lens and in the surrounding tissues, including the ganglion cell layer (Figure 2d). NMM is also robust in these tissues (Figure 2e), and Unc45b and NMM colocalize at the cortex of these cells (Figure 2l), consistent with both a role for NMM in lens development as well as association of Unc45b with NMM. At this stage, the lens is almost clear of cell nuclei (Figure 2f), but NMM and actin are still present in the retracting cells. At 3–5 dpf, the eye continues to grow and differentiate, and the lens is clear of nuclei as well as acto-myosin staining (Supplementary Figure S4).
Expression of zebrafish unc45b is detected in the lens of animals carrying a punc45b::GFP transcriptional reporter (a), as well as in fixed wild-type animals with a polyclonal zebrafish Unc45b antibody (b). Counterstain with DAPI shown in c. (d–o) Cryosections of zebrafish eye from 2 dpf embryos. Wild-type (d–f) and homozygous steif embryos (g–i) immunostained with polyclonal antibody against zebrafish Unc45b (d, g), polyclonal antibody against human Non-muscle Myosin 2A (e, h) or DAPI (f, i). Panels j and k are magnified images from panels c and d, and l is a merge of j and k, showing colocalization of Unc45b and NMM. Panels m and n are magnified images from panels g and h, and o is a merge of g and h, showing loss of cortical Unc45b staining in the mutant embryos. The steif mutation may still result in expression of a truncated, but nonfunctional protein, so polyclonal antisera still produce a signal. The transgenic line, Tg(unc45b:EGEFP)ua1, in which eGFP is driven by a 1128-bp fragment of the zebrafish unc45b promoter, was constructed at the University of Alberta by Eva Guznowski.
The unc45b mutant, steif, carries an embryonic lethal nonsense mutation in the UCS domain (Table 1 and Figure 1c).10, 20 In addition to the paralyzed phenotype, steif−/− mutants have smaller eyes compared with wild-type siblings (Figure 3). Both muscle and eye phenotypes are also seen in Unc45b dicky ticker mutants in Xenopus.21 In CC patients, the eye size is normal and none reported muscular weakness or cardial disease.
Analysis of zebrafish eye phenotypes in unc45 mutants. Lateral and dorsal views of representative 4 dpf wild type (a, a′) and steif embryos (b, b′). Steif mutants have visibly reduced ocular size and the lens does not protrude from the optic cup. Camera lucida drawing depicting the difference in head size between wild type (thin line) and steif (thick line) embryos at 48 hpf (c). Graphs showing the average eye area (d), and average pupil diameter (e) at 4 dpf and the average eye area (f) at 48 hpf. For comparison, data from a strain (kurzschluss (kus)) mutant in the second unc-45 paralog (Unc45a) as well as doubly mutant in unc45a and unc45b (kus;steif) is included.8, 26 Steif and kus;steif mutants show a significant reduction in eye area (P=0.000). Error bars indicate SEM.
At the earliest time point examined (48 hpf), the lens in steif mutants is noticeably smaller than in wild-type, and has not yet begun to clear of nuclei (Figure 2). At 3 dpf, the central lens of steif mutants contained ectopic actin fibers and nuclei that were not present in the wild-type embryos. In the wild-type lens, central lens fibers lack nuclei, and at the proximal lens pole in the lens transition zone, nuclei of the differentiating secondary fibers have a flat and elongated shape. The nuclei of steif mutants are swollen and located throughout the anterior region of the lens and the nuclei of the distal epithelium cells stain just as intensely as in wild-type embryos. In contrast to wild-type, colocalization of Unc45b and NMM is lost in steif embryos, where the presumably nonfunctional truncated protein is now seen throughout the cytoplasm (Figure 2o). Smaller eyes are often seen in zebrafish mutants lacking normal circulation, but the lens fiber disorganization at 3 dpf is not detected in the smaller eyes from pickwick mutants, defective in the heart-specific isoform of titin (Supplementary Figure S4).22 A defect in lens fiber development in steif, but not wild-type or pickwick, is also seen when eyes are stained with the ZL-1 antibody, which is fiber-cell-specific, although the nature of the antigen is unknown (Supplementary Figure S4).
RNA encoding human UNC45B was synthesized and injected into either wild-type one-cell embryos or those derived from an incross of steif heterozygous parents. Homozygous mutant embryos were determined by genotyping. Although some nuclei remain in the anterior region of the lens, their numbers appear to be reduced compared with uninjected steif embryos (Figure 4). Nuclei along the posterior edge of the lens have a flat appearance similar to those in wild-type embryos. Injection of wild-type human UNC45B mRNA was not able to rescue the ectopic F-actin localization pattern seen in steif mutants (Figure 4). RNA injections of the p.Trp805 altered form of human UNC45B into wild-type embryos did, however, recapitulate both the ectopic nuclei and F-actin staining phenotypes (Figure 4).
Cryosections of zebrafish embryonic eyes following Unc45b mRNA injections. Production of the ectopic nuclear and actin accumulation phenotypes were compared either with WT (a–d, a′–d′, m) or unc45b(steif)−/− homozygous embryos (e, f, e′, f′, p) as controls. Ectopic expression of human UNC45B in zebrafish eyes was achieved by injecting synthetic human UNC45B mRNA into one-celled embryos derived from WT or from an unc45b(steif)+/− incross. Genotype of embryos was determined retrospectively for the latter cross as described. Embryos were fixed at 3 dpf for all images. Stained with DAPI to indicate nuclei (a–p), or phalloidin to show actin localization (a′–l′). Multiple embryos are presented for each genotype. Injection of wild-type human UNC45B mRNA was able to rescue the ectopic nuclei phenotype, but not the ectopic F-actin localization pattern, seen in steif mutants (g, h, g′, h′, o). Although some nuclei remain in the anterior region of the lens, their numbers appear to be reduced compared with uninjected steif embryos. Injections of mRNA encoding the mutant form of human UNC45B into one-cell WT embryos did produce both the ectopic nuclei and F-actin staining phenotypes (i–l, i′–l′, n) reminiscent of the steif phenotype. Panels m–p show DAPI staining at higher magnification to highlight nuclei in lens.
DISCUSSION
The genome-wide linkage analysis of family CC00116 resulted in two novel CC loci on chromosomes 2 and 17, respectively.3 Additional family members were not available for exclusion of either linkage region, and targeted deep genome resequencing of the linkage regions resulted in two candidate mutations co-segregating with the disease trait. The mutations affected proteins of very different function; the ACACA gene encodes multifunctional enzyme involved in de novo biosynthesis of fatty acids and the UNC45B gene encodes a chaperone involved in correct assembly of type II muscle and NMM.
The ACACA mutation
ACACA encodes the type I fatty-acid synthase, FAS, which is expressed in the soluble cytoplasm of lipogenic tissues such as adipose, liver and mammary gland. The domain structure of FAS is conserved in eukaryotes and has evolved into the large 264 kDa synthase with seven discrete distinct functional domains.23 The p.Arg1133His mutation is in the less well-conserved central part of FAS, which has no assigned catalytic function.23 The expression of ACACA in embryonic eye tissues (Figure 1b) is consistent with a housekeeping function for de novo fatty-acid biosynthesis. Homozygous Acaca−/− knockout (KO) mouse embryos were undeveloped and died, while heterozygotes were fertile with normal life spans and normal body weight.24 Considering the function and housekeeping role of FAS and the KO mouse phenotypes, the ACACA mutation is unlikely to be the genetic cause of the CC phenotype in family CC00116. The report of the p.Arg1133His mutation as a somatic mutation in large intestine carcinoma must be considered as a passenger mutation in cancer progression.
UNC45B mutation
UNC45B is a molecular co-chaperone responsible for correct assembly of type II myosins and has a role in myoblast fusion and sarcomere organization. Within the family of UNC-45 proteins, one isoform (unc-45) is encoded in invertebrate genomes and two isoforms (Unc45a and Unc45b) in vertebrates. Unc45a is ubiquitously expressed and Unc45b is highly expressed in striated muscle25, 26 and the muscle myosin chaperone activity of UNC-45 proteins is dependent upon Hsp90 as co-chaperone.27 UNC-45 has three functionally and structurally distinct regions (Figure 1c). The N terminus contains three tetratricopeptide repeat (TPR) motifs and is involved in formation of a stable complex with the general Hsp90 protein. The central region is conserved in all metazoan UNC-45 proteins but its function still remains unclear. The C terminal consists of an UCS domain (Unc-45, Cro1 and She4,) homologous with fungal proteins involved in segregation of molecules during budding, endocytosis and cell division (Supplementary Figure S3C).25, 27, 28
Mutations are known in Caenorhabditis elegans unc-45 and in zebrafish and Xenopus tropicalis Unc45b.10, 21, 29, 30, 31 These are either lethal or temperature sensitive (ts) homozygous missense or nonsense mutations (Table 1 and Figure 1c–e). The phenotypes caused by mutations in C. elegans unc-45 or vertebrate unc45b include defective muscle development as well as effects in the vertebrate eye.10, 32 Heterozygote siblings have normal phenotypes in all systems examined. The majority of the mutations, including the human p.Arg805Trp mutation, are located in the UCS domain (Figure 1d). Lee et al.33 proposed a 3D structure from X-ray analysis of Drosophila melanogaster UNC-45, which we used to make a 3D model of the human protein (Figure 1d).34 The structure reveals a contiguous arrangement of 17 helical layers with five discrete armadillo (ARM) repeat subdomains, which is folded to an L shape with no obvious transition between the central region and the UCS domain.33 Five of the six known mutations in the UCS domain are located in a small nonpolar subregion, which has the highest degree of sequence identity in the five most evolutionarily divergent UCS proteins. This ∼40 amino-acid subregion forms the α-helices 18H1-3 and 19H1-2 (Figure 1e) and with the exception of the steif nonsense allele, all the mutations in this subdomain are missense mutations. In addition, the Arg803 position in Drosophila, which corresponds to h-Arg805 and C. elegans-Arg819, forms an inter-repeat ion pair interaction with Drosophila Glu766, which corresponds to h-Glu767 and C. elegans-Glu781.33 The C. elegans-Glu781 position is one of the ts paralyzing mutations, and the ion-interacting Arg residue is the C00116 Arg805Trp mutation. Both mutations may lead to ion pair instability of the α-helix structures (18H2 and 19H2 in Figure 1e) and reduced myosin-binding ability of the UCS domain. The heterozygous genotype of the human p.Arg805Trp mutation suggests either a dominant-negative or neomorphic mutation. This is consistent with recent evidence that the UNC45B protein function is part of a multimeric complex.35, 36 Gazda et al.36 have demonstrated that C. elegans UNC-45 forms linear protein chains that offer multiple binding sites for cooperating chaperones and client proteins. The backbone of the UNC-45 filament is composed of the tethered superhelix ARM1-8 and TPR1-3 unit, and the UCS domain extends away from the filament axis. This is consistent with the h-p.Arg805Trp alteration serving as a dominant-negative mutation that affects UNC45B multimeric complex, and suggests instability of the UNC45B polymer with structural hindrance or reduced binding stability during the assembly of nonmuscular myosin in the lens development. The occurrence of c.2413C>T heterozygous in the African American population may infer the person to have juvenile cataract. The NHLBI Exome Sequencing Project represents anonymized data, so a follow-up on the individual is not possible.
Expression and zebrafish steif mutant
UNC45B expression was seen in 43- and 54-day-old human embryonic eyes together with the two potential client proteins MYH9 and MYH10 and two co-chaperones HSP90AA1 and HSP90AB1 (Figure 1b). Expression of both Unc45b and NMM in the zebrafish embryonic eye is consistent with this analysis. Colocalization of the two proteins is seen in several different cell layers, including lens epithelia and ganglion cells, and this colocalization is disrupted in homozygous steif embryos.
The steif−/− embryos have a smaller eye compared with wild-type embryos at the same developmental stage,10 and quantitative analysis shows that the eyes were approximately half the size as seen for the wild-type or for embryos homozygous for the kurzschluss mutation, which truncates the Unc45a isoform and results in severe aortic arch defects (Figure 3).8, 26 The steif−/− embryos lens also showed persistence of nuclei compared with the wild-type embryo, suggesting the lens fiber cells maturation is affected by the unc45b mutation. The same lens phenotype showing high nuclei accumulation was seen when wild-type embryos were injected with the altered human p.Trp805 UNC45B construct (Figure 4), consistent with an ability of the mutant human UNC45B protein causing the juvenile cataract phenotype seen in family CC00116. All other unc-45/unc45b mutations described are lethal in homozygous state, where the effect of the heterozygous human p.Arg805Trp alteration results in a milder phenotype of unclear lenses. Homozygous or compound heterozygote UNC45B mutation state may be lethal in humans as seen for the zebrafish and Xenopus mutations (Table 1).
The lens grows rapidly by embryonic cell division and differentiation in the epithelial cell layer region just above the lens equator. The immature lens cells migrate below the equatorial plane into the transitional zone, where they elongate and differentiate into fiber cells.37 Data from C. elegans have shown that UNC-45 acts as chaperone in assembly of NMMs.15, 38 NMMs are known to be involved in differentiation and elongation of mouse embryo lens fiber cells where MYH10 was localized in lens epithelium and was detected at the interface of lens epithelium and fiber cells.39 The occurrence of NMM as well as its chaperone components UNC45B and HSP90 in the human and zebrafish embryo lenses suggests UNC45B may act as a chaperone in differentiation, migration and maturation of the lens epithelia cells, and the accumulation of nuclei seen in the embryo steif−/− lens (Figure 2) may be a consequence of defective maturation and organelle apoptosis in the developing lens fiber cells.
Human mutations in MYH9 (MIM 160775) are autosomal dominantly inherited40 and display phenotypically distinct disorders including May–Hegglin anomaly (MIM 155100), Fechtner syndrome (MIM 153640), and Sebastian syndrome (MIM 605249). All three disorders involve megakaryocyte, platelet, and leukocyte defects and the Fechtner syndrome is further characterized by sensorineural deafness, cataract and nephritis implicating MYH9 in the pathogenesis of cataract. Homozygous KO of Myh9 in mice was embryonic lethal, whereas heterozygote Myh9+/− mice were viable and fertile and cataract was observed in two out the three mouse lines heterozygous for human MYH9 missense mutations.16 In zebrafish, mutations that produce ocular disorders, including cataracts, are found in a similar spectrum of candidate genes as we have seen in humans, suggesting that eye development processes in the two systems are fundamentally similar at the cellular level despite differences in timing and morphology during development.4, 17, 41, 42
In conclusion, we have characterized a human UNC45B mutation in a Danish family with autosomal dominant cataract developing in early childhood. Experimental data using a zebrafish unc45b mutant show an eye developmental phenotype. Human UNC45B has been shown to be present in human embryo eye supporting a function for UNC45B in the developing lens during fiber cells maturation and migration. Both the co-chaperone HSP90 and nonmuscular myosin are expressed in the human embryo lens, which is suggestive of a role for the myosin chaperone UNC45B in lens maturation and a possible new mechanism for cataract formation.
WEB RESOURCE
1000 Genomes Browser: http://browser.1000genomes.org/index.html
AceView: http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/index.html
CatMap:2 http://cat-map.wustl.edu/
COSMIC (Catalogue of Somatic Mutations in Cancer): http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/
dbSNP, NCBI: http://www.ncbi.nlm.nih.gov/snp/
FirstGlance in Jmol: http://bioinformatics.org/firstglance/fgij/
GEPIS-tissue: http://research-public.gene.com/Research/genentech/genehub-gepis/index.html
NHLBI, Exome Sequencing Project (ESP): http://evs.gs.washington.edu/EVS/
Online Mendelian Inheritance in Man (OMIM): http://www.omim.org
Phyre234 (Protein Homology/analogY Recognition Engine V 2.0): http://www.sbg.bio.ic.ac.uk/phyre2
UCSC Genome Browser: www.genome.ucsc.edu
References
Shiels A, Hejtmancik JF : Genetic origins of cataract. Arch Ophthalmol 2007; 125: 165–173.
Shiels A, Bennett TM, Hejtmancik JF : Cat-Map: putting cataract on the map. Mol Vis 2010; 16: 2007–2015.
Hansen L, Mikkelsen A, Nurnberg P et al: Comprehensive mutational screening in a cohort of Danish families with hereditary congenital cataract. Invest Ophthalmol Vis Sci 2009; 50: 3291–3303.
Gross JM, Perkins BD : Zebrafish mutants as models for congenital ocular disorders in humans. Mol Reprod Dev 2008; 75: 547–555.
Gudbjartsson DF, Jonasson K, Frigge ML, Kong A : Allegro, a new computer program for multipoint linkage analysis. Nat Genet 2000; 25: 12–13.
Thiele H, Nurnberg P : HaploPainter: a tool for drawing pedigrees with complex haplotypes. Bioinformatics 2005; 21: 1730–1732.
Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF : Stages of embryonic development of the zebrafish. Dev Dyn 1995; 203: 253–310.
Comyn SA, Pilgrim D : Lack of developmental redundancy between Unc45 proteins in zebrafish muscle development. PLoS One 2012; 7: e48861.
Westerfield M : The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). Eugene, OR, USA: University of Oregon Press, 2000.
Etard C, Behra M, Fischer N et al: The UCS factor Steif/Unc-45b interacts with the heat shock protein Hsp90a during myofibrillogenesis. Dev Biol 2007; 308: 133–143.
Neff MM, Neff JD, Chory J, Pepper AE : dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J 1998; 14: 387–392.
Neff MM, Turk E, Kalishman M : Web-based primer design for single nucleotide polymorphism analysis. Trends Genet 2002; 18: 613–615.
Uribe RA, Gross JM : Immunohistochemistry on cryosections from embryonic and adult zebrafish eyes. CSH Protoc 2007; 2007: 4779.
French CR, Erickson T, French DV, Pilgrim DB, Waskiewicz AJ : Gdf6a is required for the initiation of dorsal-ventral retinal patterning and lens development. Dev Biol 2009; 333: 37–47.
Kachur TM, Audhya A, Pilgrim DB : UNC-45 is required for NMY-2 contractile function in early embryonic polarity establishment and germline cellularization in C. elegans. Dev Biol 2008; 314: 287–299.
Zhang Y, Conti MA, Malide D et al: Mouse models of MYH9-related disease: mutations in nonmuscle myosin II-A. Blood 2012; 119: 238–250.
Greiling TMS, Clark JI : Early lens development in the zebrafish: a three-dimensional time-lapse analysis. Dev Dyn 2009; 238: 2254–2265.
Greiling TMS, Aose M, Clark JI : Cell fate and differentiation of the developing ocular lens. Invest Ophthalmol Vis Sci 2010; 51: 1540–1546.
Weber GF, Menko AS : Actin filament organization regulates the induction of lens cell differentiation and survival. Dev Biol 2006; 295: 714–729.
Wohlgemuth SL, Crawford BD, Pilgrim DB : The myosin co-chaperone UNC-45 is required for skeletal and cardiac muscle function in zebrafish. Dev Biol 2007; 303: 483–492.
Geach TJ, Zimmerman LB : Paralysis and delayed Z-disc formation in the Xenopus tropicalis unc45b mutant dicky ticker. BMC Dev Biol 2010; 10: 75.
Xu X, Meiler SE, Zhong TP et al: Cardiomyopathy in zebrafish due to mutation in an alternatively spliced exon of titin. Nat Genet 2002; 30: 205–209.
Rangan V, Smith S : Fatty acid synthesis in eukaryotes; in Vance D, Vance J (eds): Biochemistry of lipids, lipoproteins and membranes. Amsterdam, Netherlands: Elsevier Science BV, 2002.
Maier T, Leibundgut M, Boehringer D, Ban N : Structure and function of eukaryotic fatty acid synthases. Q Rev Biophys 2010; 43: 373–422.
Price MG, Landsverk ML, Barral JM, Epstein HF : Two mammalian UNC-45 isoforms are related to distinct cytoskeletal and muscle-specific functions. J Cell Sci 2002; 115: 4013–4023.
Anderson MJ, Pham VN, Vogel AM, Weinstein BM, Roman BL : Loss of unc45a precipitates arteriovenous shunting in the aortic arches. Dev Biol 2008; 318: 258–267.
Barral JM, Hutagalung AH, Brinker A, Hartl FU, Epstein HF : Role of the myosin assembly protein UNC-45 as a molecular chaperone for myosin. Science 2002; 295: 669–671.
Etheridge L, DiIorio P, Sagerstrom CG : A zebrafish unc-45-related gene expressed during muscle development. Dev Dyn 2002; 224: 457–460.
Epstein HF, Thomson JN : Temperature-sensitive mutation affecting myofilament assembly in C. elegans. Nature 1974; 250: 579–580.
Barral JM, Bauer CC, Ortiz I, Epstein HF : Unc-45 mutations in Caenorhabditis elegans implicate a CRO1/She4p-like domain in myosin assembly. J Cell Biol 1998; 143: 1215–1225.
Venolia L, Ao W, Kim S, Kim C, Pilgrim D : unc-45 gene of Caenorhabditis elegans encodes a muscle-specific tetratricopeptide repeat-containing protein. Cell Motil Cytoskeleton 1999; 42: 163–177.
Ao W, Pilgrim D : Caenorhabditis elegans UNC-45 is a component of muscle thick filaments and colocalizes with myosin heavy chain B, but not myosin heavy chain A. J Cell Biology 2000; 148: 375–384.
Lee CF, Hauenstein AV, Fleming JK et al: X-ray crystal structure of the UCS domain-containing UNC-45 myosin chaperone from Drosophila melanogaster. Structure 2011; 19: 397–408.
Kelley LA, Sternberg MJE : Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 2009; 4: 363–371.
Shi H, Blobel G : UNC-45/CRO1/She4p (UCS) protein forms elongated dimer and joins two myosin heads near their actin binding region. Proc Natl Acad Sci USA 2010.
Gazda L, Pokrzywa W, Hellerschmied D et al: The myosin chaperone UNC-45 is organized in tandem modules to support myofilament formation in C. elegans. Cell 2013; 152: 183–195.
Lovicu FJ, McAvoy JW : Growth factor regulation of lens development. Dev Biol 2005; 280: 1–14.
Kachur T, Ao W, Berger J, Pilgrim D : Maternal UNC-45 is involved in cytokinesis and co-localizes with a non-muscle myosin in the early Caenorhabditis elegans embryo. J Cell Science 2004; 117: 5313–5321.
Maddala R, Skiba N, Vasantha Rao P : Lens fiber cell elongation and differentiation is associated with a robust increase in myosin light chain phosphorylation in the developing mouse. Differentiation 2007; 75: 713–725.
Seri M, Pecci A, Di Bari F et al: MYH9-related disease: May-Hegglin anomaly, Sebastian syndrome, Fechtner syndrome, and Epstein syndrome are not distinct entities but represent a variable expression of a single illness. Medicine 2003; 82: 203–215.
Goishi K, Shimizu A, Najarro G et al: AlphaA-crystallin expression prevents gamma-crystallin insolubility and cataract formation in the zebrafish cloche mutant lens. Development 2006; 133: 2585–2593.
Greiling TMS, Houck SA, Clark JI : The zebrafish lens proteome during development and aging. Mol Vis 2009; 15: 2313–2325.
Acknowledgements
We are grateful to the family for their participation. Lars Allan Larsen is thanked for donation of the human embryo RNA, Annemette Mikkelsen, Linda Boje Dalsgaard and Karen Friis Henriksen are thanked for library preparation, cDNA synthesis and expression analyses. The cataract study was supported by a grant from The Danish Association of the Blind, Wilhelm Johannsen Centre for Functional Genome Research hosted the targeted next generation sequencing, and RCLink hosted the RNA expression analyses. Research in the Pilgrim lab is supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (NSERC). We are grateful to Andrew Waskiewicz for the use of his zebrafish facility, which is supported by the NSERC MRS Program. SC was supported by a Graduate Studentship from NSERC.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Hansen, L., Comyn, S., Mang, Y. et al. The myosin chaperone UNC45B is involved in lens development and autosomal dominant juvenile cataract. Eur J Hum Genet 22, 1290–1297 (2014). https://doi.org/10.1038/ejhg.2014.21
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2014.21
Keywords
This article is cited by
-
Bi-allelic mutations in uncoordinated mutant number-45 myosin chaperone B are a cause for congenital myopathy
Acta Neuropathologica Communications (2019)
-
Genetic landscape of isolated pediatric cataracts: extreme heterogeneity and variable inheritance patterns within genes
Human Genetics (2019)
-
The zebrafish eye—a paradigm for investigating human ocular genetics
Eye (2017)
-
Melanosomes in pigmented epithelia maintain eye lens transparency during zebrafish embryonic development
Scientific Reports (2016)
-
Loss of function of myosin chaperones triggers Hsf1-mediated transcriptional response in skeletal muscle cells
Genome Biology (2015)