Abstract
Here we characterized the genome-wide architecture of copy number variations (CNVs) in 286 healthy, unrelated Finnish individuals belonging to the MUSGEN study, where molecular background underlying musical aptitude and related traits are studied. By using Illumina HumanOmniExpress-12v.1.0 beadchip, we identified 5493 CNVs that were spread across 467 different cytogenetic regions, spanning a total size of 287.83 Mb (∼9.6% of the human genome). Merging the overlapping CNVs across samples resulted in 999 discrete copy number variable regions (CNVRs), of which ∼6.9% were putatively novel. The average number of CNVs per person was 20, whereas the average size of CNV per locus was 52.39 kb. Large CNVs (>1 Mb) were present in 4% of the samples. The proportion of homozygous deletions in this data set (∼12.4%) seemed to be higher when compared with three other populations. Interestingly, several CNVRs were significantly enriched in this sample set, whereas several others were totally depleted. For example, a CNVR at chr2p22.1 intersecting GALM was more common in this population (P=3.3706 × 10−44) than in African and other European populations. The enriched CNVRs, however, showed no significant association with music-related phenotypes. Moreover, the most common CNV locations in world’s normal population cohorts (6q14.1, 11q11) were overrepresented in this population. Thus, the genome-wide CNV investigation in this Finnish sample set demonstrated features that are characteristic to isolated populations. Novel CNVRs and the functional implications of CNVs revealed in this study elucidate structural variation present in this population isolate, and may also serve as candidate gene loci for music-related traits.
Similar content being viewed by others
Introduction
Copy number variation (CNV) represents the major portion of variation in the human genome with respect to size1, 2, 3, 4 and is known for its role in altering gene expression, thereby affecting genetic diversity, evolution and disease risk.5, 6, 7 The evaluation of the role of a CNV in disease risk relies on its frequency in normal population cohorts.8, 9 Many such cohorts exhibited inter- and intra-population differences in CNV frequency distributions.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22 In addition to disease risk, these differences, furthermore, explain a significant proportion of normal phenotypic variation.23, 24, 25, 26
In this context, we characterized the genome-wide architecture of CNVs in 286 healthy, unrelated subjects characterized for musical aptitude and related traits.27 We wanted to essentially evaluate the role of CNV enrichment in music-related phenotypes. In a broader perspective, the sample set represents the isolated Finnish population that has experienced multiple bottlenecks in its population history.28 Owing to founder effect and genetic drift, 36 monogenic disorders (caused by one major mutation) were enriched in this population, whereas many other rare monogenic disorders such as phenylketonuria and maple syrup disease were depleted.28, 29 Moreover, CNV remains poorly characterized in genetically isolated populations, including the Finnish population. Characterization of the genome-wide architecture of CNVs in this sample set thus enables the genotype–phenotype correlation, and provides novel insights into normal structural variation of a population isolate.
Methods
Study material
The study material comprised 286 healthy, unrelated Finnish subjects (167 females, 119 males; mean age of 55.21 years; range 18–94 years) who participate in the MUSGEN project, where molecular background of musical aptitude and related traits are studied.27 The participants neither reported any relatives in the study (based on a questionnaire) nor showed any close relatedness in identity by descent analysis. No medical information was available from the participants, but as far as we know, they are healthy. The Ethical Committee of Helsinki University Central Hospital approved this study. An informed consent was obtained from all participants. More information about the MUSGEN project and the sample recruitment can be found in the Supplementary information.
Genotyping
For genotyping, we used 200 ng of DNA that was extracted from the peripheral blood of each subject (no cell lines were used). All the samples were genotyped using Illumina Infinium HumanOmniExpress-12v1.0 beadchip (730 K; San Diego, CA, USA), with an average overall call rate of 99.54%. Normalized signal intensity data was obtained through Illumina BeadStudio software. Normalized measures of total signal intensity (Log2 R ratios) and the relative allelic signal intensity ratio (B-allele frequencies) at each marker were used for CNV identification in all samples.
CNV detection
CNVs were identified using two algorithms: PennCNV30 and QuantiSNP,31 and only the consistent calls were retained for further analyses. All probe coordinates in this study were mapped to human genome build GRCh37/hg19. We followed two different approaches for constructing a CNV map, which are: (1) so-called copy number variable region (CNVR) using any-overlap criterion4, 8, 11, 12, 13, 16, 17, 18, 20, 21 and (2) copy number variable cytogenetic region (CNVcR) representing any cytogenetic region that contains one or more CNVs. The study protocol is shown in Supplementary Figure S1. Detailed descriptions of all the methods with their associated references are provided in the methods section of Supplementary material.
Results
General characteristics of CNVs
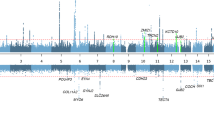
We observed a total of 5493 CNV events in 267 samples that passed the quality control evaluation. Of these, 3888 (70.7%) CNV events were deletions, whereas 1605 (29.3%) were duplications. Notably, 12.4% of the autosomal CNVs were homozygous deletions, whereas only 0.05% constituted four-copy duplication. On average, ∼20 CNV events (14 deletions and 6 duplications) were discovered per person. We observed at least one CNV of size >100 kb in almost 90% of the samples and of size >500 kb in ∼13% of samples, where 4% of the samples had a CNV >1 Mb (Supplementary Table S1). The total CNV size accounted for 287.83 Mb (147.67 Mb deletions and 140.15 Mb duplications), whereas the average size of CNV per locus was 52.39 kb (37.98 kb for deletions and 87.32 kb for duplications). Approximately 75% of the total CNV events were <50 kb and ∼40% of them were <10 kb. (Figure 1, Supplementary Table S2). A recent study32 has provided evidence for age-related accumulation of CNVs. In our sample, we did not find statistically significant difference in the numbers of CNVs, large CNVs, novel CNVs between elderly (>60 years of age; N=107) and middle-aged (<55 years of age; N=133) subjects. Unfortunately, longitudinal follow-up of the appearance of CNVs was not available in this study.
Genome-wide map of CNVs
We followed two different criteria for the construction of a genome-wide CNV map in this sample set. From the 5493 consistent CNV events, a total of 999 CNVRs (618 (61.9%) deletions, 381 (38.1%) duplications) were constructed. Among the 999 CNVRs, 631 (63.2%) regions contained rare CNVs (<1% population), whereas 368 (36.8%) regions contained polymorphic CNVs2, 3(>1% population).
A total of 467 different cytogenetic regions contained CNVs in this sample set. We define such cytogenetic regions as CNVcRs in this study. Of these 467 CNVcRs, 190 regions (40.68%) were found to be rare (<1% population), whereas 277 regions (59.31%) were polymorphic (>1% of population). We tested whether the highly frequent CNVcRs in this study (Table 1) were significantly overrepresented or underrepresented compared with other populations. For this, we computed the frequencies of CNVcRs from 39 studies representing 9793 samples. Specifically, we combined the CNV data from the database of genomic variants1 (DGV; 37 studies, 8528 samples); Vogler et al16 (1167 samples) and Teo et al18 (98 samples). Excepting 14q11.2, four other highly frequent CNVcRs (6q14.1, 11q11, 2p22.3, 3q28) were found to be significantly overrepresented among the Finns (two-sided Fisher’s exact test; P-value (fdr) <0.05) (Table 1).
Novel CNVR characteristics
We found that ∼6.9% of the CNVRs detected in this study were novel, whereas ∼93.1% were already known and cataloged in DGV. Nearly 83% of the novel CNVRs in our data were rare (<1% population). Approximately 93% of the novel CNVRs were <50 kb. Although ∼50% of the novel CNVRs overlapped with Refseq genes, only half of them overlapped exonic regions. Notably, homozygous deletions were not observed in the novel CNVRs.
We further compared the CNVRs in this study with CNV calls from few individual studies to assess the level of concordance. Highest concordance was observed with the study of Shaikh et al,14 whose study was based on Illumina Infinium II Human-Hap550 beadchip with 65% of their sample set comprising Caucasians. Overall, depending on the CNV detection methodology (platforms and algorithms) and ethnic background of the population, the level of concordance with different studies varied significantly (Supplementary Table S3).
Enrichment of CNVs and their phenotype–genotype correlation
We checked if any particular CNV was significantly enriched or depleted in this sample set (two-sided Fisher’s exact test, P-value threshold: 0.05) compared with (1) mixed Caucasian and African–Americans,14 (2) African and Swiss populations16 and (3) Swedish population.18 Interestingly, several CNVRs showed significant enrichment and were observed only in the Finnish sample set (Table 2a). In fact, some of the enriched CNVRs intersected genes that affect brain function. For example, CNVRs overlapping protocadherin alpha gene cluster (PCHDA1-9; 47 subjects), glucose mutarotase gene (GALM; 45 subjects) and cGMP-dependent protein kinase type I (PRKG1; 23 subjects) are notably relevant for brain function and could be relevant candidates for musical traits. Several other common CNVRs in Finnish sample set such as chr8:51031221-51040022 containing SNTG1 (P=1.7 × 10−37) and chr16:28615243-28620752 with SULT1A1 (P=1.2 × 10−31) were not reported in other populations. In this connection, analysis of the music-related phenotypes (COMB scores and creativity in music; detailed in Supplementary information) among the enriched CNV carriers showed no significant excess of the phenotypes in either carriers or noncarriers (data not shown).
On the other hand, several CNVRs that were relatively common in other populations were not observed in the Finnish population (Table 2b). Putative functions of the genes intersected by these enriched and depleted CNVRs are shown in Supplementary Table S5.
Genomic impact of CNVs in the finnish sample set
A total of 491 (49.1%) CNVRs overlapped with 835 RefSeq genes of which 321 genes (38.4%) were deleted, whereas 423 genes (50.7%) were duplicated. In all, 91 genes (10.9%) have undergone both deletions and duplications, whereas 37 genes (4.4%) were overlapping with novel CNVRs. Table 3 shows some of the clinically important CNVs and genes that were significantly polymorphic in the population.
The most common CNVR contained genes from the olfactory gene cluster (OR4C11, OR4P4, OR4S2), amylase gene cluster (AMY1A, AMY1B, AMY1C) and protocadherin alpha gene cluster (PCDHA1-9) (Supplementary Figure S2). Of the 835 Refseq genes that fell within CNVs, 396 genes were present in the OMIM database, which contains information on all Mendelian disorders, whereas 593 genes were present in PharmGKB, the pharmacogenomics knowledge base (more functional categories detailed in Supplementary Table S4).
To identify the enriched functional categories falling within CNVRs of this study, we used a hypergeometric distribution test implemented in Genetrail.33 A stringent P-value threshold (fdr) of 0.01 resulted in the enrichment of only one KEGG pathway; olfactory transduction (P=2.07 × 10−6). In addition to this, several gene ontology terms were significantly enriched in our CNV data set (P-value (fdr); threshold 0.01) (Supplementary Figure S3). These enriched functional categories included: (1) biological processes such as cell-adhesion, sensory perception, cognition and neurological system process, (2) cellular component terms pertaining to the membrane parts and (3) molecular function terms such as molecular transducer activity, alpha-amylase activity and olfactory receptor activity.
Discussion
This study presents the first comprehensive CNV map of 286 healthy, unrelated subjects belonging to MUSGEN project, who originate from the isolated Finnish population. Primarily, this genome-wide CNV investigation in the Finnish sample set demonstrated features that are characteristic to isolated populations. In particular, highly significant enrichment of certain CNVRs and a total depletion of other CNVRs in this population suggest a founder effect. Adding strength to this finding, even the most common CNV locations in the world’s normal population cohorts (6q14.1, 11q11) were overrepresented in this population (Table 1). In addition, CNVs in this population comprised a higher proportion of homozygous deletions than three other populations from a recent study,19 hinting at a founder effect.
Further, ∼6.9% of the CNVRs detected in this population sample was novel. The majority of those novel CNVRs were small (<50 kb), suggesting that smaller variants in the human genome have not been comprehensively characterized yet. Although the functional impact of such smaller variants has often been underestimated, recent studies34, 35 have shown that smaller variants (often intergenic) affect transcription factor binding and, consequently, gene expression. Moreover, identification of ∼93% known variants indicates that the detection methodology used in this study was sufficiently sensitive to capture known variations.
The enriched CNVRs-intersecting genes such as PCDHA1-9, GALM and PRKG1 are intriguing because of their remarkable relevance for brain function. PCHDA1-9 gene cluster is related to the serotonergic systems that influences neurocognitive and motor functions,36 and was found to be cosegregated with low-music test scores in a recent family-based CNV study.37 GALM is associated with serotonin transporter binding potential in the human thalamus,38 whereas PRKG1, expressed in the neurons of amygdala, was suggested to support synaptic plasticity.39 Although these CNVRs showed no statistically significant association with the music-related phenotypes, owing to a limited sample size, we cannot exclude their role as possible candidate genes for musical aptitude and related traits.
Several CNVs detected in this study have considerable clinical relevance. Specifically, some of the highly polymorphic (> 5% population) CNVs intersected known disease-related genes (Table 3), which may intrigue the public health sector. Lack of disease markers screening in the study participants, makes it unfeasible to exclude the probability of an identified CNV to be potentially predisposing for disease conditions. In addition, 13% of the individuals in this study accommodated at least one large, rare CNV. In this regard, it is worth noting that these large CNVs are typically rare in normal population cohorts,12 but have a potential role in neuropsychiatric diseases.5, 40, 41, 42 Further investigations are warranted but remain beyond the scope of this study.
The functional impact of common CNVs in the Finnish population appeared to be consistent with the world’s populations in a number of aspects. Firstly, duplications overlapped more genes than deletions in our study, supporting the presumption of deletions being biased away from genes.4, 43 Secondly, our findings aligned with the idea of rare CNVs being large and harboring more genes than common events.12 Further, significantly enriched gene ontology terms in the Finnish CNVs included extracellular biological processes, such as sensory perception, cognition and neurological system process that are in accordance with the findings of previous population-based CNV studies.4, 15, 17, 22, 43 In fact, the genes affecting these biological processes are also intriguing for music-related phenotypes. Also, a recent CNV study in swine species44 reported similarity in the enriched functional categories that allows us to speculate that CNVs are comparable across different species. Focusing on individual genes, we found that genes from the olfactory gene cluster (OR4C11, OR4P4, OR4S2), amylase gene cluster (AMY1A, AMY1B, AMY1C) and protocadherin alpha gene cluster (PCDHA1-9) were relatively more frequent among the CNVs in this population. Regardless of their frequencies in the Finnish population, these genes have been widely described in CNVs in previous studies.45, 46, 47
The general characteristics of CNVs detected in this study are similar to those in previous studies that used the same technology platform. For instance, the number of CNVs, deletion/duplication ratio and the average size of CNV (20, 2.4:1 and 52.39 kb, respectively) in this study are comparable to the statistics of Shaikh et al.14 and Xu et al.19 Both of these studies used an Illumina SNP array with relatively less marker density. Moreover, when we estimated the degree of overlap between Finnish CNVs and CNVs from 10 different studies, we found a higher degree of overlap with studies based on Illumina SNP array.
In a broader perspective, array-based CNV studies often involve several discrepancies in detection, cataloging and comparisons. Most importantly, differences in the array architecture, choice of algorithms, population differences and phenotypes affect our understanding of CNVs.48, 49, 50, 51 Being aware of all such discrepancies, we chose to abide by widely adopted methods to make our data comparable across studies. However, an increase in the sample size and a higher marker density in this study would fine-tune the estimates of population frequencies and their correlation with the studied phenotypes.
In conclusion, this first-generation CNV map of the MUSGEN project originating from the isolated Finnish population shows features characteristic to a founder effect. It would be interesting to see whether a similar phenomenon can be detected in other population isolates. The enriched CNVRs and biological processes suggest pathways important in evolution52 and may serve as candidate genes for music-related traits. Finally, these findings help future studies on music-related phenotypes, human genetic diseases, and demographic history, as well as contribute to the global variation map of CNVs.
References
Iafrate AJ, Feuk L, Rivera MN et al: Detection of large-scale variation in the human genome. Nat Genet 2004; 36: 949–951.
Sebat J, Lakshmi B, Troge J et al: Large-scale copy number polymorphism in the human genome. Science 2004; 305: 525–528.
Feuk L, Carson AR, Scherer SW : Structural variation in the human genome. Nat Rev Genet 2006; 7: 85–97.
Redon R, Ishikawa S, Fitch KR et al: Global variation in copy number in the human genome. Nature 2006; 444: 444–454.
Lupski JR : Genomic rearrangements and sporadic disease. Nat Genet 2007; 39: S43–S47.
Henrichsen CN, Chaignat E, Reymond A : Copy number variants, diseases and gene expression. Hum Mol Genet 2009; 18: R1–R8.
Zhang F, Gu W, Hurles ME, Lupski JR : Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet 2009; 10: 451–481.
Scherer SW, Lee C, Birney E et al: Challenges and standards in integrating surveys of structural variation. Nat Genet 2007; 39: S7–S15.
Eichler EE, Flint J, Gibson G et al: Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet 2010; 11: 446–450.
White SJ, Vissers LE, Geurts van Kessel A et al: Variation of CNV distribution in five different ethnic populations. Cytogenet Genome Res 2007; 118: 19–30.
Zogopoulos G, Ha KC, Naqib F et al: Germ-line DNA copy number variation frequencies in a large North American population. Hum Genet 2007; 122: 345–353.
Itsara A, Cooper GM, Baker C et al: Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet 2009; 84: 148–161.
Li J, Yang T, Wang L et al: Whole genome distribution and ethnic differentiation of copy number variation in Caucasian and Asian populations. PLoS ONE 2009; 4: e7958.
Shaikh TH, Gai X, Perin JC et al: High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res 2009; 19: 1682–1690.
Park H, Kim JI, Ju YS et al: Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing. Nat Genet 2010; 42: 400–405.
Vogler C, Gschwind L, Rothlisberger B et al: Microarray-based maps of copy-number variant regions in European and sub-Saharan populations. PLoS ONE 2010; 5: e15246.
Yim SH, Kim TM, Hu HJ et al: Copy number variations in East-Asian population and their evolutionary and functional implications. Hum Mol Genet 2010; 19: 1001–1008.
Teo SM, Ku CS, Naidoo N et al: A population-based study of copy number variants and regions of homozygosity in healthy Swedish individuals. J Hum Genet 2011; 56: 524–533.
Xu H, Poh WT, Sim X et al: SgD-CNV, a database for common and rare copy number variants in three Asian populations. Hum Mutat 2011; 32: 1341–1349.
Lou H, Li S, Yang Y et al: A map of copy number variations in Chinese populations. PLoS ONE 2011; 6: e27341.
Wineinger NE, Pajewski NM, Kennedy RE et al: Characterization of autosomal copy-number variation in African Americans: the HyperGEN Study. Eur J Hum Genet 2011; 19: 1271–1275.
Moon S, Kim YJ, Hong CB, Kim DJ, Lee JY, Kim BJ : Data-driven approach to detect common copy-number variations and frequency profiles in a population-based Korean cohort. Eur J Hum Genet 2011; 19: 1167–1172.
Yeo RA, Gangestad SW, Liu J, Calhoun VD, Hutchison KE : Rare copy number deletions predict individual variation in intelligence. PLoS ONE 2011; 6: e16339.
Dauber A, Yu Y, Turchin MC et al: Genome-wide association of copy-number variation reveals an association between short stature and the presence of low-frequency genomic deletions. Am J Hum Genet 2011; 89: 751–759.
Kim YK, Moon S, Hwang MY et al: Gene-based copy number variation study reveals a microdeletion at 12q24 that influences height in the Korean population. Genomics 2013; 101: 134–138.
Macleod AK, Davies G, Payton A et al: Genetic copy number variation and general cognitive ability. PLoS ONE 2012; 7: e37385.
Pulli K, Karma K, Norio R, Sistonen P, Goring HH, Jarvela I : Genome-wide linkage scan for loci of musical aptitude in Finnish families: evidence for a major locus at 4q22. J Med Genet 2008; 45: 451–456.
Peltonen L, Jalanko A, Varilo T : Molecular genetics of the Finnish disease heritage. Hum Mol Genet 1999; 8: 1913–1923.
Norio R : Finnish Disease Heritage II: population prehistory and genetic roots of Finns. Hum Genet 2003; 112: 457–469.
Wang K, Li M, Hadley D et al: PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 2007; 17: 1665–1674.
Colella S, Yau C, Taylor JM et al: QuantiSNP: an objective Bayes hidden-Markov model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res 2007; 35: 2013–2025.
Forsberg LA, Rasi C, Razzaghian HR et al: Age-related somatic structural changes in the nuclear genome of human blood cells. Am J Hum Genet 2012; 90: 217–228.
Backes C, Keller A, Kuentzer J et al: GeneTrail--advanced gene set enrichment analysis. Nucleic Acids Res 2007; 35: W186–W192.
Kasowski M, Grubert F, Heffelfinger C et al: Variation in transcription factor binding among humans. Science 2010; 328: 232–235.
Schlattl A, Anders S, Waszak SM, Huber W, Korbel JO : Relating CNVs to transcriptome data at fine resolution: assessment of the effect of variant size, type, and overlap with functional regions. Genome Res 2011; 21: 2004–2013.
Katori S, Hamada S, Noguchi Y et al: Protocadherin-alpha family is required for serotonergic projections to appropriately innervate target brain areas. J Neurosci 2009; 29: 9137–9147.
Ukkola-Vuoti L, Kanduri C, Oikkonen J et al: Genome-wide copy number variation analysis in extended families and unrelated individuals characterized for musical aptitude and creativity in music. PLoS ONE 2013; 8: e56356., doi:10.1371/journal.pone.0056356.
Liu X, Cannon DM, Akula N et al: A non-synonymous polymorphism in galactose mutarotase (GALM) is associated with serotonin transporter binding potential in the human thalamus: results of a genome-wide association study. Mol Psychiatry 2011; 16: 584–585.
Paul C, Schoberl F, Weinmeister P et al: Signaling through cGMP-dependent protein kinase I in the amygdala is critical for auditory-cued fear memory and long-term potentiation. J Neurosci 2008; 28: 14202–14212.
Stefansson H, Rujescu D, Cichon S et al: Large recurrent microdeletions associated with schizophrenia. Nature 2008; 455: 232–236.
Pinto D, Pagnamenta AT, Klei L et al: Functional impact of global rare copy number variation in autism spectrum disorders. Nature 2010; 466: 368–372.
Cooper GM, Coe BP, Girirajan S et al: A copy number variation morbidity map of developmental delay. Nat Genet 2011; 43: 838–846.
Conrad DF, Pinto D, Redon R et al: Origins and functional impact of copy number variation in the human genome. Nature 2010; 464: 704–712.
Wang J, Jiang J, Fu W et al: A genome-wide detection of copy number variations using SNP genotyping arrays in swine. BMC Genomics 2012; 13: 273.
Noonan JP, Grimwood J, Schmutz J, Dickson M, Myers RM : Gene conversion and the evolution of protocadherin gene cluster diversity. Genome Res 2004; 14: 354–366.
Perry GH, Dominy NJ, Claw KG et al: Diet and the evolution of human amylase gene copy number variation. Nat Genet 2007; 39: 1256–1260.
Young JM, Endicott RM, Parghi SS, Walker M, Kidd JM, Trask BJ : Extensive copy-number variation of the human olfactory receptor gene family. Am J Hum Genet 2008; 83: 228–242.
Winchester L, Yau C, Ragoussis J : Comparing CNV detection methods for SNP arrays. Brief Funct Genomic Proteomic 2009; 8: 353–366.
Dellinger AE, Saw SM, Goh LK, Seielstad M, Young TL, Li YJ : Comparative analyses of seven algorithms for copy number variant identification from single nucleotide polymorphism arrays. Nucleic Acids Res 2010; 38: e105.
Pinto D, Darvishi K, Shi X et al: Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat Biotechnol 2011; 29: 512–520.
Haraksingh RR, Abyzov A, Gerstein M, Urban AE, Snyder M : Genome-wide mapping of copy number variation in humans: comparative analysis of high resolution array platforms. PLoS ONE 2011; 6: e27859.
Poptsova M, Banerjee S, Gokcumen O, Rubin MA, Demichelis F : Impact of constitutional copy number variants on biological pathway evolution. BMC Evol Biol 2013; 13: 19.
Acknowledgements
The Academy of Finland (grant reference no. 13371), The Biocentrum Helsinki Foundation and the Wellcome Trust (grant reference no. 090532/Z/09/Z) supported this work. We thank all the participants of this study for their generous cooperation. We are grateful to Minna Varhala for expert technical help. We also gratefully acknowledge the contribution of CNV data from previous studies that were used in this study for comparative analyses.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Kanduri, C., Ukkola-Vuoti, L., Oikkonen, J. et al. The genome-wide landscape of copy number variations in the MUSGEN study provides evidence for a founder effect in the isolated Finnish population. Eur J Hum Genet 21, 1411–1416 (2013). https://doi.org/10.1038/ejhg.2013.60
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2013.60
Keywords
This article is cited by
-
Population history modulates the fitness effects of Copy Number Variation in the Roma
Human Genetics (2023)
-
How far musicality and perfect pitch are derived from genetic factors?
Journal of Applied Genetics (2020)
-
Convergent evidence for the molecular basis of musical traits
Scientific Reports (2016)
-
Compilation of copy number variants identified in phenotypically normal and parous Japanese women
Journal of Human Genetics (2014)