Abstract
Cystic fibrosis (CF) is caused by mutations in the CF transmembrane conductance regulator gene (CFTR). Disease severity in CF varies greatly, and sibling studies strongly indicate that genes other than CFTR modify disease outcome. Syntaxin 1A (STX1A) has been reported as a negative regulator of CFTR and other ion channels. We hypothesized that STX1A variants act as a CF modifier by influencing the remaining function of mutated CFTR. We identified STX1A variants by genomic resequencing patients from the Bernese CF Patient Data Registry and applied linear mixed model analysis to establish genotype–phenotype correlations, revealing STX1A rs4363087 (c.467−38A>G) to significantly influence lung function. The same STX1A risk allele was recognized in the European CF Twin and Sibling Study (P=0.0027), demonstrating that the genotype–phenotype association of STX1A to CF disease severity is robust enough to allow replication in two independent CF populations. rs4363087 is in linkage disequilibrium to the exonic variant rs2228607 (c.204C>T). Considering that neither rs4363087 nor rs2228607 changes the amino-acid sequence of STX1A, we investigated their effects on mRNA level. We show that rs2228607 reinforces aberrant splicing of STX1A mRNA, leading to nonsense-mediated mRNA decay. In conclusion, we demonstrate the clinical relevance of STX1A variants in CF, and evidence the functional relevance of STX1A variant rs2228607 at molecular level. Our findings show that genes interacting with CFTR can modify CF disease progression.
Similar content being viewed by others
Introduction
Cystic fibrosis (CF) is the most common severe autosomal-recessive disorder in Caucasians and is characterized by a progressive decline in lung function due to an exaggerated and sustained inflammatory response to pathogens, as well as by malabsoprtion due to pancreatic insufficiency.1 The disease is caused by mutations of the CF transmembrane conductance regulator gene (CFTR), which functions as an apical chloride channel and negative regulator of other ion channels like the amiloride-sensitive sodium channel (ENaC) in airway and intestinal epithelial membranes. The pathophysiology of the disease has not yet been fully elucidated, yet recent studies indicate that the absence of negative regulation of ENaC might be as crucial as the absence of chloride transport by CFTR in CF epithelia. Anomalies in ion flux would then lead to a volume-depleted periciliary liquid, resulting in increased mucus viscosity and impaired cough clearance.2
The phenotypic expression of CF shows a broad variability, even among patients carrying the same CFTR genotype. Besides environmental and other factors, variability is due to modifier genes3 that influence the immune response in CF4, 5, 6 or modify ion flux at the apical membrane.7 In addition, it has been proposed that members of the so-called CFTR-interactome influence CF clinics.8, 9
Syntaxin 1A (brain) (STX1A) is a t-SNARE protein involved in the fusion of synaptic vesicles and neurotransmitter release. Previous studies have shown that STX1A is expressed in airway and intestinal cells,10 where it negatively modulates CFTR function by altering intracellular trafficking and/ or channel activity.11 In addition, STX1A was found to negatively regulate the remaining function of the most common CFTR mutation, p.Phe508del.10 STX1A consists of 288 amino-acid residues, of which the 23 C-terminal residues serve as membrane anchor. The isoform syntaxin 1C (STX1C) contains an insert of 91 bp at codon 226, leading to a frameshift and subsequently to a truncated protein of 260 residues lacking the transmembrane domain. STX1C is expressed in several tissues, yet it is unclear whether it is present in the lung.12 In the following, the term ‘STX1A/1C’ is used for all analyses and statements that do not explicitly distinguish between the two isoforms.
In the current study, we hypothesized that CF disease outcome might be influenced by STX1A/C variants that either increase or hamper STX1A/C functionality leading to a further reduction or enhancement of the remaining CFTR function by the regulatory activity of STX1A/C. Modeling lung function data collected from the Bernese CF cohort with linear-mixed models revealed that STX1A/C variants significantly influence CF disease progression, a finding that could be confirmed in the European CF Twin and Sibling Study cohort. One of the STX1A/C variants, rs2228607, was shown to modulate the efficiency of an aberrant splice event triggering nonsense-mediated mRNA decay (NMD), indicating that this variant is functionally relevant. Intriguingly, expression studies evidenced strong expression of STX1C, raising the question which of the two isoforms may in fact mediate the effect of the variant.
Materials and methods
A detailed version of materials and methods can be found in Supplementary Data.
Patients
From the Bernese CF Patient Data Registry,13 56 CF patients homozygous for CFTR p.Phe508del and with biometric and lung function data from age 6 to 20 years were analyzed (see Supplementary Data for detailed materials and methods). Lung function data included lung clearance index (LCI), forced expiratory volume in 1 s (FEV1), forced expiratory flow (FEF) at 50% (FEF50) and specific airway resistance (sReff), as well as the functional residual capacity and volume of trapped gas .
For replication of the association study, the independent cohort of the European CF Twin and Sibling Study was analyzed.14 Evaluation of STX1A/C genotype data was based on transmitted chromosomes of the entire patient population of 101 CF families (171 p.Phe508del-CFTR homozygotes) and the following subpopulations selected for extreme clinical phenotypes: concordant mildly affected patient pair (CON+ 13 families), concordant severely affected patient pair (CON− 12 families) and discordant sib pairs (DIS; 14 families).14 Informed consent was obtained from all subjects, and the local ethics committees approved the study.
STX1A/C genotyping
The 10 exons and the adjacent intronic regions of the STX1A/C gene were screened for variants in the Bernese cohort by SSCP/ HD analysis. In the patients from the European Twin and Sibling Study, STX1A/C SNPs rs2228607 and rs4363087 were genotyped using restriction enzyme-based assays.
Statistics
In the Bernese cohort, linear mixed models were used to assess the relationship between the repeated measurements of each lung function parameter and STX1A/C variants.9 Models were calculated with PROC MIXED of SAS 9.2. In order to correct for multiple testing (six lung function parameters in three genotypes) and to reduce the risk of false positives, only results with P<0.002 were considered statistically significant. In the European Sib and Twin Study cohort, genetic data of the association study were evaluated using the FAMHAP software package, which accepts data evaluation in association studies on unrelated individuals as well as on affected sib pairs.15 The raw observed P values (Praw) were corrected for multiple testing by haplotype permutation (Pglobal).16
STX1A/C transcript characterization
Nasal epithelial cells were obtained from CF patients and healthy individuals as previously described.17 For NMD inhibition, the epithelial cells were incubated in culture medium supplemented with 10 mM caffeine for 6.5 h at 37 °C prior to lysis.18, 19 Extraction and reverse transcription of total RNA was followed by PCR amplification and sequencing of STX1A/C exons 1–6 and 5–10. ARMS-PCR20 either amplifying the T or the C allele at rs2228607 was performed in individuals heterozygous for rs2228607 after caffeine treatment. Proportions of PCR products corresponding either to correct splicing or to partial inclusion of intron 3 were determined by fragment analysis. To assess expression ratios of STX1A and STX1C, isoform-specific qPCR was performed as previously described.21 The effect of rs2228607 on splicing was additionally assessed with the in-silico tool Human Splicing Finder (www.umd.be/HSF).
Results
Influence of STX1A/C variants on clinical outcome
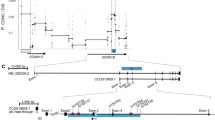
Screening of the 10 STX1A/C exons and the adjacent intronic regions in 56 Bernese CF patients revealed two synonymous and eight non-coding variants, which were all in Hardy–Weinberg equilibrium (Table 1). Analyzing the impact on lung function of the three variants with a frequency >10% (rs2228607, rs4363087, and rs35459363) revealed a significantly worse progression of FEV1, sReff, and LCI in patients carrying the G allele at rs4363087 (P<0.002 in linear mixed models; Supplementary Table S1). FEV1 (z-score) of patients homozygous for the G allele at rs4363087 decreased by 0.2340 while LCI increased by 0.6488 per year as compared to patients homozygous for the A allele. Heterozygous patients showed an intermediate disease progression as compared to homozygotes (Figure 1). rs4363087 was additionally found to be in linkage disequilibrium with rs2228607, with the T allele at rs2228607 being associated with the G allele at rs4363087.
Results of linear mixed models, represented by regressions derived from coefficients of variant rs4363087. Solid line, rs4363087 AA; dotted line, rs4363087 AG, thick line, rs4363087 GG. CF patients carrying the G allele at rs4363087 (STX1A/C c.467−38G) show significantly worse lung disease progression than patients homozygous for the A allele.
Three non-overlapping subsamples of the European CF twin and sib pairs, stratified for the pair’s extreme clinical phenotype, were compared at the informative STX1A/C markers rs2228607 and rs4363087 (Table 2), which displayed a minor allele frequency >0.4 in the patient population of 171 p.Phe508del-CFTR homozygous European CF twins and siblings. STX1A/C diplotype distributions of concordant severely affected pairs (CON−) differed significantly from those of concordant mildly affected pairs (CON+ Pglobal=0.0385; corrected for multiple testing, Table 2) and from those of discordant patient pairs (DIS; Pglobal=0.0027; corrected for multiple testing, Table 2). Homozygosity for the T allele at rs2228607 and homozygosity for the G allele at rs4363087 was overrepresented among CON− pairs, whereby these genotypes were observed on more than 50% of CON− patients (Table 2). Homozygosity for the resulting rs2228607–rs4363087 diplotype T–G was not observed among DIS patients, observed on less than a fifth of CON+ patients but on more than half of the CON− siblings (Table 2). We conclude that the risk allele at the STX1A/C locus resides on the rs2228607–rs4363087 T–G haplotype, replicating the finding among the Bernese cohort that disease severity as measured by LCI and FEV1 progression is associated with the G allele at rs4363087. Furthermore, the overrepresentation of T–G homozygotes among the CON− sample indicates that the STX1A/C risk allele is rs2228607– rs4363087 T–G recessive.
STX1A/C transcript characterization and expression studies
As neither rs4363087 nor rs2228607 being in linkage disequilibrium with it changes the amino-acid sequence of STX1A/C, we analyzed STX1A/C mRNA in order to reveal a potential mode of action of these variants.
In a first step, we sequenced cDNA amplicons of nasal epithelia spanning exons STX1A/C 1–6 and 5–10, respectively, which evidenced that the 3′ splice site of intron 8 was shifted 91 bp upstream as compared to the STX1A refseq (NM_004603.3). This splice isoform corresponds to the previously described STX1A isoform STX1C.15
In order to confirm absent or low STX1A mRNA expression in nasal epithelium, we performed splice-isoform-specific qPCR of STX1A and STX1C, revealing that STX1C mRNA is >350 times higher expressed than STX1A. Similar results were obtained in all epithelia investigated, with STX1C being at least 50 times higher expressed than STX1A (Figure 2). In order to test the capability of our assay to properly detect STX1A, we additionally applied it to IMR-32 cells expressing high levels of STX1A,22 and indeed high levels of STX1A were demonstrated in these cells (Figure 2).
Expression ratios of STX1C and STX1A as assessed with isoform-specific qPCR. Expression ratios were calculated using the formula 2^[Cq(STX1A)−Cq(STX1C)]. STX1C showed higher expression in all epithelia investigated, while STX1A was highly expressed in IMR-32 neuronal cells. Details on the investigated cell lines are supplied in Supplementary Data.
We next treated nasal epithelium samples with caffeine in order to identify aberrant STX1A/C splice variants prone to NMD. Qualitative transcript analysis revealed a 151-bp inclusion of intron 3 (r.208_209ins209-539_209-389, r.1 being the adenosine of the start codon of NM_004603.3) appearing upon caffeine treatment. This partial intron inclusion leads to a premature stop codon (p.Lys70SerfsX3) and is thus an NMD substrate.
As rs2228607 is located in proximity of the 5′ splice site of intron 3, we next evaluated if this variant modifies the frequency of the observed intron 3 inclusion. Of the 13 samples investigated, 12 showed more intron 3 inclusion with the C allele at rs2228607 than with the T allele (median: 2.1 times more intron 3 inclusion with allele C than T; range 1.3–4.4 times). One patient showed no intron 3 inclusion associated with the C allele; however, he was the only one of the 13 patients carrying concomitantly the variant c.150C>T in a heterozygous state.
Finally, in silico analysis of the effect of rs2228607 on splicing indicated that the T allele of rs2228607 disrupts a binding site of the splice factor SC35.
Discussion
In the current study, we investigated whether or not variants of the well-characterized CFTR interactor STX1A modify CF disease outcome.
As the same STX1A/C allele carrying G at rs4363087 (c467.38A>G) was recognized as a risk allele in the two non-overlapping CF patient populations from Bernese (P<0.002; Supplementary Table S1) and from the European CF Twin and Sibling Study (P=0.0027; Table 2), we concur that our observed association of STX1A/C genotype to CF disease severity is robustly replicated in two independent CF patient populations and by two complementary strategies of analysis, that is, genotype–phenotype association (Bernese study) and an association study comparing case and reference populations (European CF Twin and Sibling Study). The latter evidenced an odds ratio of 3.5 for rs4363087 alleles in association with severely affected sibs (Table 2). Allele-specific amplification of rs2228607, being in linkage disequilibrium with rs4363087, revealed the C allele at rs2228607 (c.204C) to be associated with increased aberrant splicing of intron 3, presumably enhancing NMD-based degradation of STX1A/C mRNA.
Previous studies aiming at identifying so-called CF modifiers mainly focused on genes of the immune system and on ion channels other than CFTR. However, many modifiers identified in single, small populations by candidate gene approaches could not be reproduced and/or lacked functional analyses of the investigated variants. Among the best established CF lung disease modifiers are mannose binding lectin,23 interferon-related developmental regulator 1,5 and transforming growth factor-beta1, with the risk genotype of the latter having an odds ratio of 2.2 in association with severe lung disease.4 As CF modifiers identified so far could not fully explain the phenotypic variability observed in CF, whole-genome association studies were initiated in order to detect yet unknown pathways associated with CF severity.24
We now show that members of the CFTR-interactome modify CF disease outcome. Importantly, STX1A/C presumably exerts its effect by modifying the functionality of CFTR. This suggests that besides immunological factors and ion channels other than CFTR, genetic variants of factors associated with trafficking and regulation of CFTR and other ion channels might additionally be key players in CF pathophysiology. This finding does not only provide new targets for candidate gene studies, but will also facilitate evaluation of whole-genome association studies. Moreover, our findings open the possibility that STX1A variants act as natural CFTR potentiators, disclosing new therapeutic approaches in CF.
As none of the variants associated with rs4363087 changes the amino-acid sequence of STX1A/C, mRNA analyses were performed, indicating that STX1A/C rs2228607 is associated with aberrant splicing of intron 3 and NMD. This aberrant splicing might be associated with reduced binding of the splice factor SC35. Reduced stability of STX1A/C mRNA carrying the C allele at rs2228607 could explain the phenotypic features associated with this variant; however, we cannot exclude that rs4363087 or other variants being in LD with these variants might have additional functional consequences. Accordingly, further investigations are required to fully elucidate the impact of these variants on STX1A/C function. Hypothetically, STX1A/C variants could exert their effect on the CF phenotype as follows: presence of the variant reduces the functionality of the isoform STX1A, for example, by enhancing aberrant splicing and thus NMD. As STX1A has a negative impact on CFTR function,25 decreased STX1A functionality would enhance the remaining function of CFTR p.Phe508del, resulting in a less severe CF phenotype.
Comparison of allele frequencies in concordant and discordant sib pairs additionally indicated the presence of a transmodulator7 interacting with the A allele at rs4363087 and still increasing the complexity of the relationship between STX1A/C and CF disease. Basically, this finding is in line with reports demonstrating CF disease modulation by complex interactions, for example, between distinct gene variants as well as the environment,6, 23 and suggests that CF phenotypes originate from a complex interplay of a multitude of factors.
Intriguingly, our STX1A/C expression studies in various epithelial cells demonstrate high expression of STX1C. Evidence of high STX1C expression is in contrast to previous studies reporting solely expression of STX1A in epithelia,10 a discrepancy that can be explained by the fact that these studies used an antibody not discriminating between STX1A and STX1C. Regrettably, in the literature only STX1A–CFTR interactions have been investigated and described so far, while potential STX1C–CFTR interactions have been neglected probably due to the fact that STX1C expression has mainly aroused interest in the research community in the context of its neuronal expression. Thus, it is still unclear which of the two variant isoforms exert their effect on CF phenotypes.
In summary, we present both clinical and functional evidence that STX1A/C variants modify CF disease outcome, demonstrating the importance of CFTR interactors in CF pathophysiology.
References
Ratjen F, Döring G : Cystic fibrosis. Lancet 2003; 361: 681–689.
Mall MA : Role of the amiloride-sensitive epithelial Na+ channel in the pathogenesis and as a therapeutic target for cystic fibrosis lung disease. Exp Physiol 2009; 94: 171–174.
Knowles MR, Drumm M : The influence of genetics on cystic fibrosis phenotypes. Cold Spring Harb Perspect Med 2012; 2: a009548.
Drumm ML, Konstan MW, Schluchter MD et al: Genetic modifiers of lung disease in cystic fibrosis. N Engl J Med 2005; 353: 1443–1453.
Gu Y, Harley ITW, Henderson LB et al: Identification of IFRD1 as a modifier gene for cystic fibrosis lung disease. Nature 2009; 458: 1039–1042.
Collaco JM, Vanscoy L, Bremer L et al: Interactions between secondhand smoke and genes that affect cystic fibrosis lung disease. JAMA 2008; 299: 417–424.
Stanke F, Becker T, Cuppens H et al: The TNFalpha receptor TNFRSF1A and genes encoding the amiloride-sensitive sodium channel ENaC as modulators in cystic fibrosis. Hum Genet 2006; 119: 331–343.
Wang X, Venable J, LaPointe P et al: Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell 2006; 127: 803–815.
Gisler FM, von Kanel T, Kraemer R, Schaller A, Gallati S : Identification of SNPs in the cystic fibrosis interactome influencing pulmonary progression in cystic fibrosis. Eur J Hum Genet 2013; 21: 397–403.
Naren AP, Di A, Cormet-Boyaka E et al: Syntaxin 1A is expressed in airway epithelial cells, where it modulates CFTR Cl(-) currents. J Clin Invest 2000; 105: 377–386.
Naren AP, Nelson DJ, Xie W et al: Regulation of CFTR chloride channels by syntaxin and Munc18 isoforms. Nature 1997; 390: 302–305.
Jagadish MN, Tellam JT, Macaulay SL, Gough KH, James DE, Ward CW : Novel isoform of syntaxin 1 is expressed in mammalian cells. Biochem J 1997; 321: 151–156.
Kraemer R, Baldwin DN, Ammann RA, Frey U, Gallati S : Progression of pulmonary hyperinflation and trapped gas associated with genetic and environmental factors in children with cystic fibrosis. Respir Res 2006; 7: 138.
Stanke F, Becker T, Kumar V et al: Genes that determine immunology and inflammation modify the basic defect of impaired ion conductance in cystic fibrosis epithelia. J Med Genet 2011; 48: 24–31.
Herold C, Becker T : Genetic association analysis with FAMHAP: a major program update. Bioinformatics 2009; 25: 134–136.
Knapp M, Becker T : Family-based association analysis with tightly linked markers. Hum Hered 2003; 56: 2–9.
Steiner B, Truninger K, Sanz J, Schaller A, Gallati S : The role of common single-nucleotide polymorphisms on exon 9 and exon 12 skipping in nonmutated CFTR alleles. Hum Mutat 2004; 24: 120–129.
Ivanov I, Lo KC, Hawthorn L, Cowell JK, Ionov Y : Identifying candidate colon cancer tumor suppressor genes using inhibition of nonsense-mediated mRNA decay in colon cancer cells. Oncogene 2007; 26: 2873–2884.
Dormer RL, Dérand R, McNeilly CM et al: Correction of delF508-CFTR activity with benzo(c)quinolizinium compounds through facilitation of its processing in cystic fibrosis airway cells. J Cell Sci 2001; 114: 4073–4081.
Newton CR, Graham A, Heptinstall LE et al: Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res 1989; 17: 2503–2516.
Amaral MD, Clarke LA, Ramalho AS et al: Quantitative methods for the analysis of CFTR transcripts/splicing variants. J Cyst Fibros 2004; 3 (Suppl 2): 17–23.
Nakayama T, Mikoshiba K, Yamamori T, Akagawa K : Expression of syntaxin 1C, an alternative splice variant of HPC-1/syntaxin 1A, is enhanced by phorbol-ester stimulation in astroglioma: participation of the PKC signaling pathway. FEBS Lett 2003; 536: 209–214.
Dorfman R, Sandford A, Taylor C et al: Complex two-gene modulation of lung disease severity in children with cystic fibrosis. J Clin Invest 2008; 118: 1040–1049.
Wright FA, Strug LJ, Doshi VK et al: Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat Genet 2011; 43: 539–546.
Naren AP, Quick MW, Collawn JF, Nelson DJ, Kirk KL : Syntaxin 1A inhibits CFTR chloride channels by means of domain-specific protein-protein interactions. Proc Natl Acad Sci USA 1998; 95: 10972–10977.
Acknowledgements
We are thankful to the patients and clinicians involved in this study, in particular Dr Martin Schöni and Dr Carmen Casaulta. The Swiss National Foundation, Grants No. 3200-066767.01 and 310000-112652, supported this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
von Kanel, T., Stanke, F., Weber, M. et al. Clinical and molecular characterization of the potential CF disease modifier syntaxin 1A. Eur J Hum Genet 21, 1462–1466 (2013). https://doi.org/10.1038/ejhg.2013.57
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2013.57
Keywords
This article is cited by
-
Exocytosis-related genes and response to methylphenidate treatment in adults with ADHD
Molecular Psychiatry (2018)
-
Pluripotent stem cells for disease modeling and drug screening: new perspectives for treatment of cystic fibrosis?
Molecular and Cellular Pediatrics (2015)