Abstract
Apolipoprotein E, encoded by APOE, is the main apoprotein for catabolism of chylomicrons and very low density lipoprotein. Two common single-nucleotide polymorphisms (SNPs) in APOE, rs429358 and rs7412, determine the three epsilon alleles that are established genetic risk factors for late-onset Alzheimer’s disease (AD), cerebral amyloid angiopathy, and intracerebral hemorrhage (ICH). These two SNPs are not present in most commercially available genome-wide genotyping arrays and cannot be inferred through imputation using HapMap reference panels. Therefore, these SNPs are often separately genotyped. Introduction of reference panels compiled from the 1000 Genomes project has made imputation of these variants possible. We compared the directly genotyped and imputed SNPs that define the APOE epsilon alleles to determine the accuracy of imputation for inference of unobserved epsilon alleles. We utilized genome-wide genotype data obtained from two cohorts of ICH and AD constituting subjects of European ancestry. Our data suggest that imputation is highly accurate, yields an acceptable proportion of missing data that is non-differentially distributed across case and control groups, and generates comparable results to genotyped data for hypothesis testing. Further, we explored the effect of imputation algorithm parameters and demonstrated that customization of these parameters yields an improved balance between accuracy and missing data for inferred genotypes.
Similar content being viewed by others
Introduction
Apolipoprotein E (APOE) is an essential mediator for catabolism of chylomicrons and very low density lipoprotein remnants. There are three major APOE isoforms, APOE2, APOE3, and APOE4, which differ in amino acids 112 and 158, determined by single-nucleotide polymorphisms (SNPs) rs429358 and rs7412, respectively.1 These variants collectively constitute the epsilon (ɛ) alleles ɛ2, ɛ3, and ɛ4, corresponding to the three human APOE isoforms. The ɛ4 allele is robustly associated with increased risk and decreased age of onset of Alzheimer’s disease (AD), whereas ɛ2 has a protective effect.2, 3, 4, 5 These alleles have also been implicated in other neurological and non-neurological disorders, including cerebral amyloid angiopathy, lobar intracerebral hemorrhage (ICH), and hyperlipidemia.6, 7 However, the absence of these SNPs from most genome-wide genotyping platforms, coupled with the inability to impute them using HapMap-based reference panels have precluded evaluation of their possible role in other diseases in the context of genome-wide association studies. The advent of comprehensive reference panels based on the 1000 Genomes project has allowed imputation of the two variants in GWA data. In fact, this approach has already been used in association studies examining the epsilon alleles.8 However, the accuracy of imputation and the distribution of missing data obtained using this approach have not been systematically evaluated. In this study, we assessed the accuracy of the 1000-Genome-based imputation for inferring unobserved epsilon allele-defining SNPs, evaluated the distribution of missing data after imputation across case and control groups, and compared association testing in directly genotyped and imputed variants.
Materials and methods
This analysis utilized data drawn from studies of ICH and AD. The ICH data set comprised individuals of European ancestry recruited in the Genetics of Cerebral Hemorrhage with Anticoagulation (GOCHA) study, a multicenter prospective cohort study of primary ICH.9 Control subjects were randomly selected from the same population using a clinic-based sampling technique. Subjects with ICH were classified as lobar when the hematoma originated in the cerebral cortico–subcortical junction, or non-lobar ICH when the hemorrhage was located in deep supratentorial structures or in infratentorial locations.9 The AD cohort consisted of individuals from the Alzheimer's disease neuroimaging initiative (ADNI), a longitudinal study of individuals with mild cognitive impairment and early AD, as well as cognitively normal older individuals.10 Both studies were approved by the institutional review board and ethics committees of participating institutions, and written informed consent was obtained from all participants or their next of kin.
For direct genotyping of the epsilon allele-defining variants in GOCHA, DNA was extracted from blood, quantified using the Quant-iT Broad-Range DNA Assay Kit (Invitrogen, Life Technologies, Carlsbad, CA, USA), and normalized to the concentration of 30 ng/μl. rs429358 and rs7412 were genotyped in two separate assays using the TaqMan SNP Genotyping Assay (Life Technologies), and the epsilon alleles were determined; the T allele at both SNPs identifies the ɛ2 allele, whereas the C allele at both positions constitute the ɛ4 allele. The T allele at rs429358 and the C allele at rs7412 identify the ɛ3 allele, which is the most common epsilon allele in general population. In ADNI, direct genotyping was performed by PCR amplification, digestion of PCR products using the HhaI restriction enzyme, and resolution of fragments on 4% MetaPhor agarose gel.
Genome-wide genotyping was performed in both groups using Illumina HumanHap610 quad array (San Diego, CA, USA) and variants were called by BeadStudio v3.2. Genome-wide genotyping data of subjects enrolled in GOCHA have been deposited in the database of genotypes and phenotypes (http://tinyurl.com/qj5exm2). Quality control of the genome-wide data was performed and samples with the following criteria were excluded: genotype call rate <95%, genome-wide heterozygosity >34.5 or <31.5 (±3 SDs from the mean), discordant clinical and genotypic gender, and pi-hat>0.1875.11 Principal component analysis was performed incorporating genotypes from Phase 3 HapMap populations. The majority of subjects clustered with the CEU (Northern Europeans from Utah) and TSI (Tuscans from Italy) HapMap populations. Population outliers were identified and removed by visual inspection of principal component plots. SNP quality control filters were genotyping rate <95%, minor allele frequency (MAF) <1%, case-control differential missingness, and departure from the Hardy–Weinberg equilibrium calculated in the entire data at P <1E-06.
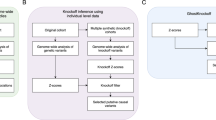
Subsequently, IMPUTE2 v2.3.0 was used to impute unobserved SNPs based on the 1000-Genome Phase I (Interim, release date June 2011) reference panel.12, 13 Imputation was initially completed using default parameters (K parameter=80, iteration number=30) and the standard threshold of 0.9 for hard-calling the dosages for the epsilon allele-defining SNPs. In order to evaluate the impact of imputation parameters and hard-calling threshold on the accuracy and missingness rate, imputation was performed using a wide range of hard-calling threshold, as well as two parameters of the imputation algorithm, namely K parameter and number of iterations. These parameters are key options that control the Markov chain Monte Carlo (MCMC) algorithm used by IMPUTE2 program; the K parameter determines the number of haplotypes used as templates for phasing the observed genotypes. The total number of the MCMC algorithm iterations is controlled by the iteration number option. Increasing these values is expected to improve imputation accuracy but at the cost of longer analysis times. We also assessed the accuracy of imputation in pre-phased genotypes generated using SHAPEIT v1.14
Agreement between imputed and genotyped SNPs was assessed by Cohen’s kappa coefficient, and differential missingness across cases and controls was evaluated using the χ2-test. Logistic regression was utilized for association testing, assuming additive genetic effects separately for the ɛ2 and ɛ4 alleles (1degree-of-freedom trend test), and adjusting for age, sex and principal components. Hypothesis testing involved the Wald test performed on the regression parameters of each epsilon allele. Quality control, principal component analysis, and association testing were performed using PLINK v1.07 and R version 2.15.2.15
Results
After quality control procedures and principal component analysis, 327 case and 250 control subjects in the GOCHA cohort, and 407 case and 202 control subjects in the ADNI cohort were available for analysis (Supplementary Table 1). As expected, the ɛ3 allele was the most common allele in case and control subjects combined, with frequency of 76% and 65% in GOCHA and ADNI, respectively. Using the default imputation parameters and hard-calling threshold of 0.9, we were able to infer rs429358 in 88% and rs7412 in 90% of subjects in GOCHA. In the ADNI cohort, these variants were ascertained in 81% and 86% of individuals, respectively. Similar to direct genotyping, the imputation of rs429358 seems to be less efficient compared with rs7412. In fact, the missingness of rs429358 was higher compared with rs7412 in both GOCHA and ADNI, whereas it was statistically significant only in ADNI (P=0.056 vs P=0.008). The rate of missing genotype for none of the SNPs was significantly different between case and control groups in both cohorts (P>0.1). A high degree of correlation between imputed and genotyped SNPs was observed in GOCHA with kappa values of 0.94 for rs429358 and 0.93 for rs7412. In ADNI, kappa coefficients were 0.92 and 0.9 for the two variants, respectively (Table 1).
The results of imputation using customized parameters suggest that the parameter K is inversely associated with the rate of missing genotypes, but its effect on kappa is less consistent (Figure 1 and Supplementary Figure 1). The iteration number of 100 yielded the best results for both variants consistent across both cohorts. Applying the default imputation parameters with the hard-calling threshold of 0.8 reduced the missing rate from about 13–14% to 7–9% in GOCHA, whereas its effect on correlation was relatively small (0.93 vs 0.91). The rate of missing genotypes and kappa coefficient changed to a similar degree when testing in ADNI. Evaluating the imputation in the pre-phased data with the default hard-calling threshold, we observed reduction in the missing rate to 5–9% in the two cohorts, but kappa impaired (ranging between 0.81 and 0.89).
Efficiency and accuracy of imputation of APOE epsilon allele-defining SNPs in the intracerebral hemorrhage cohort. (a, b) Correlation coefficient between genotyped and imputed rs429358 and rs7412 across a range of K parameter, iteration number, and hard-call threshold values. The corresponding missing rates are plotted in the bottom panels. K, K parameter.
Association testing yielded similar effect estimates and P-values for the genotyped and imputed alleles across both cohorts (Table 2). Though underpowered to detect the known effects of the ɛ2 and ɛ4 alleles in ICH (40% and 62% power, respectively), the results for the ɛ4 allele are compatible with previous reports.6 The association testing in the AD cohort demonstrated increased risk of AD in individuals carrying the ɛ4 allele. The odds ratio for the genotyped ɛ4 was 4 and 3.51 for the imputed allele, with the P-value of 7.62E-16 and 7.12E-10, respectively.
Discussion
The APOE epsilon alleles have a potent role in the risk of several complex diseases and have been implicated in an extraordinary range of additional disorders.16 Despite the accumulation of genome-wide array data for many of these phenotypes, it has been difficult to confirm the effect of epsilon alleles because of limitations in the coverage of array designs. Most of the genome-wide genotyping arrays that have been widely used in GWA studies so far do not include rs429358 and rs7412, owing to relatively higher failure of genotyping, especially for rs429358, and limited contribution of these SNPs to the imputation of the entire locus, which has a complex linkage disequilibrium structure. In addition, direct genotyping of these SNPs may not be feasible owing to logistical issues such as inadequate DNA samples, or because of increase in time and costs. Our analysis demonstrates that the epsilon allele-defining variants can be imputed successfully by taking advantage of the reference panel based on the 1000 Genomes project. Imputation can be performed with high accuracy, an acceptable proportion of missing data, and absence of differential missingness in inferred genotypes across case and control groups. This provides the opportunity for complementary analysis on currently available GWA data without the need to perform direct genotyping. Studies have already begun to implement imputation to infer epsilon alleles and it is expected that further studies will be performed using this approach.
Customization of imputation parameters and hard-call threshold can yield a lower proportion of missing data without significant decrease in accuracy. Although a proportion of genotypes are missed with imputation, causing variable decreases in power, this is not expected to yield false-positive results owing to information bias as the missing genotypes are evenly distributed across case and control groups. Nevertheless, it remains crucial to ensure that the missing genotypes are symmetrically distributed across the study groups before proceeding to association testing, especially when analyzing data obtained from subjects with relatively higher frequency of the risk alleles.
We used the 1000-Genome Phase I Interim reference panel. It is demonstrated that imputation performance improves with the latest release, Phase I integrated haplotypes. However, the gain in imputation performance is mainly observed for SNPs with MAF<5%, and particularly those with MAF<2%, providing only a marginal impact in this particular imputation scenario.17 Although this study was performed in two relatively small data sets, similar results were obtained. Further analyses employing larger samples could provide broader insight into this topic.
References
Laws SM, Hone E, Gandy S, Martins RN : Expanding the association between the APOE gene and the risk of Alzheimer's disease: possible roles for APOE promoter polymorphisms and alterations in APOE transcription. J Neurochem 2003; 84: 1215–1236.
Corder EH, Saunders AM, Strittmatter WJ et al: Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 1993; 261: 921–923.
Pastor P, Roe CM, Villegas A et al: Apolipoprotein Eepsilon4 modifies Alzheimer's disease onset in an E280A PS1 kindred. Ann Neurol 2003; 54: 163–169.
Saunders AM, Strittmatter WJ, Schmechel D et al: Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer's disease. Neurology 1993; 43: 1467–1472.
West HL, Rebeck GW, Hyman BT : Frequency of the apolipoprotein E epsilon 2 allele is diminished in sporadic Alzheimer disease. Neurosci Lett 1994; 175: 46–48.
Biffi A, Sonni A, Anderson CD et al: Variants at APOE influence risk of deep and lobar intracerebral hemorrhage. Ann Neurol 2010; 68: 934–943.
Donnelly LA, Palmer CN, Whitley AL et al: Apolipoprotein E genotypes are associated with lipid-lowering responses to statin treatment in diabetes: a Go-DARTS study. Pharmacogenet Genomics 2008; 18: 279–287.
Lill CM, Liu T, Schjeide BM et al: Closing the case of APOE in multiple sclerosis: no association with disease risk in over 29 000 subjects. J Med Genet 2012; 49: 558–562.
Genes for Cerebral Hemorrhage on Anticoagulation Collaborative G: Exploiting common genetic variation to make anticoagulation safer. Stroke 2009; 40: S64–S66.
Mueller SG, Weiner MW, Thal LJ et al: Ways toward an early diagnosis in Alzheimer's disease: the Alzheimer's disease neuroimaging initiative (ADNI). Alzheimers Dement 2005; 1: 55–66.
Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT : Data quality control in genetic case-control association studies. Nat Protoc 2010; 5: 1564–1573.
Genomes Project C, Abecasis GR, Altshuler D et al: A map of human genome variation from population-scale sequencing. Nature 2010; 467: 1061–1073.
Howie BN, Donnelly P, Marchini J : A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009; 5: e1000529.
Delaneau O, Marchini J, Zagury JF : A linear complexity phasing method for thousands of genomes. Nature Methods 2012; 9: 179–181.
Purcell S, Neale B, Todd-Brown K et al: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Verghese PB, Castellano JM, Holtzman DM : Apoliporotein E in Alzheimer's disease and other neurological disorders. Lancet Neurol 2011; 10: 241–252.
Delaneau O, Marchini J : The 1000 Genomes Project Consortium. Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Under review 2013, Available at http://mathgen.stats.ox.ac.uk/impute/data_download_1000G_phase1_integrated_SHAPEIT2.html.
Acknowledgements
The Genetics of Cerebral Hemorrhage with Anticoagulation study was funded by NIH-NINDS grant R01NS059727, the Keane Stroke Genetics Research Fund, the Edward and Maybeth Sonn Research Fund, by the University of Michigan General Clinical Research Center (M01 RR000042), and by a grant from the National Center for Research Resources. GJF was supported by the NIH-NINDS SPOTRIAS fellowship grant P50NS061343. CDA was supported by a Clinical Research Training Fellowship from the American Brain Foundation. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; BioClinica, Inc; Biogen Idec Inc; Bristol-Myers Squibb Company; Eisai Inc; Elan Pharmaceuticals, Inc; Eli Lilly and Company; F Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc; GE Healthcare; Innogenetics, NV; IXICO Ltd; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC; Medpace, Inc; Merck & Co, Inc; Meso Scale Diagnostics, LLC; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc; Piramal Imaging; Servier; Synarc Inc; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, Rev October 16, 2012 San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129 and K01 AG030514. The investigators within the ADNI contributed to the design and implementation of the ADNI and/or provided data, but did not participate in analysis or writing of this report.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Radmanesh, F., Devan, W., Anderson, C. et al. Accuracy of imputation to infer unobserved APOE epsilon alleles in genome-wide genotyping data. Eur J Hum Genet 22, 1239–1242 (2014). https://doi.org/10.1038/ejhg.2013.308
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2013.308
Keywords
This article is cited by
-
Frailty and the risk of dementia: is the association explained by shared environmental and genetic factors?
BMC Medicine (2021)
-
Considering the APOE locus in Alzheimer’s disease polygenic scores in the Health and Retirement Study: a longitudinal panel study
BMC Medical Genomics (2020)
-
Common germline variants of the human APOE gene modulate melanoma progression and survival
Nature Medicine (2020)
-
Effect of BDNF Val66Met on hippocampal subfields volumes and compensatory interaction with APOE-ε4 in middle-age cognitively unimpaired individuals from the ALFA study
Brain Structure and Function (2020)
-
Apolipoprotein E polymorphism and the risk of aneurysmal subarachnoid hemorrhage in a South Indian population
Cellular & Molecular Biology Letters (2017)