Abstract
Hereditary diffuse gastric cancer (HDGC) syndrome, although rare, is highly penetrant at an early age, and is severe and incurable because of ineffective screening tools and therapy. Approximately 45% of HDGC families carry germline CDH1/E-cadherin alterations, 20% of which are nonsense leading to premature protein truncation. Prophylactic gastrectomy is the only recommended approach for all asymptomatic CDH1 mutation carriers. Suppressor-tRNAs can replace premature stop codons (PTCs) with a cognate amino acid, inducing readthrough and generating full-length proteins. The use of suppressor-tRNAs in HDGC patients could therefore constitute a less invasive therapeutic option for nonsense mutation carriers, delaying the development of gastric cancer. Our analysis revealed that 23/108 (21.3%) of E-cadherin-mutant families carried nonsense mutations that could be potentially corrected by eight suppressor-tRNAs, and arginine was the most frequently affected amino acid. Using site-directed mutagenesis, we developed an arginine suppressor-tRNA vector to correct one HDGC nonsense mutation. E-cadherin- deficient cell lines were transfected with plasmids carrying simultaneously the suppressor-tRNA and wild-type or mutant CDH1 mini-genes. RT-PCR, western blot, immunofluorescence, flow cytometry and proximity ligation assay (PLA) were used to establish the model, and monitor mRNA and protein expression and function recovery from CDH1 vectors. Cells expressing a CDH1 mini-gene, carrying a nonsense mutation and the suppressor-tRNA, recovered full-length E-cadherin expression and its correct localization and incorporation into the adhesion complex. This is the first demonstration of functional recovery of a mutated causative gene in hereditary cancer by cognate amino acid replacement with suppressor-tRNAs. Of the HDGC families, 21.3% are candidates for correction with suppressor-tRNAs to potentially delay cancer onset.
Similar content being viewed by others
Introduction
Hereditary diffuse gastric cancer (HDGC; OMIM #137215) is an autosomal dominant disorder and accounts for 1% of all gastric cancer (GC) cases.1, 2 Although rare, HDGC is incurable, severe, highly penetrant, presents at early age and lacks effective screening tools and therapy. The main cause of cancer lethality in HDGC families is the development of an aggressive type of cancer where noncohesive neoplastic cells infiltrate the normal epithelia, generating diffuse gastric cancer (DGC) or, less frequently, lobular breast cancer (LBC). HDGC is mainly associated with inherited defects in CDH1 (which encodes for the protein E-cadherin (ENSG00000039068; OMIM #192090)). Germline heterozygous point or small frameshift mutations and large deletions explain ∼45% of all HDGC families.3, 4 Notably, germline CDH1 mutations may also be rarely found in families with LBC, lacking mutations in breast cancer-associated genes,5 and in patients without prior descriptions of cancer familial aggregation, but presenting early onset of DGC (EODGC).6
Approximately 80% of germline CDH1 mutations are truncating,7, 8, 9 20% of which are nonsense.10 Missense mutations, present in ∼20% of HDGC families, may be highly deleterious.2 CDH1 germline mutation carriers have an 80% risk of developing DGC, and up to 60% risk of developing LBC (in females), at the age of 80 years.1 Somatic inactivation of the remaining CDH1 wild-type (WT) allele, in cells from HDGC patient’s stomach or breast, determines cancer initiation, often in the second or third decades of life.11, 12 The resulting undetected preclinic multifocal tumours have increased spreading ability, leading to diagnosis at advanced stages, high risk of relapse after surgery and poor prognosis.1, 11 Multimodal therapy regimens have limited success, making presymptomatic diagnosis detrimental.9 The only currently available strategy to manage asymptomatic CDH1 mutation carriers is total prophylactic gastrectomy that, despite the associated morbidity, prevents disease development at least in stomach.1, 13 Female CDH1 mutation carriers are recommended to undergo annual breast cancer surveillance, starting at the age of 35 years, but may also choose for prophylactic mastectomy.14 Importantly, a nonneglectable proportion of patients refuse prophylactic interventions.
For all the above-mentioned reasons, E-cadherin-related diseases, globally referred to as HDGC, constitute a good model for testing nonsurgical prophylactic strategies. In the current report, we propose using nonsense suppression, as a way of reducing cancer risk in CDH1 germline mutation carriers, to the risk observed for general population.
Important developments have been made recently in the field of nonsense suppression applied to inherited syndromes such as Duchenne muscular dystrophy or cystic fibrosis, especially using aminoglycosides15 and PTC124.16 However, these agents may introduce missense mutations that may be detrimental in cancer syndromes, by affecting protein structure or function.17, 18, 19 In contrast, suppressor-tRNAs (sup-tRNAs) are able to induce stop codon readthrough by specifically introducing the cognate amino acid at the premature stop codon (PTC) site.20 As each sup-tRNA can recognize and interact with one of the three stop codons amber (UAG), opal (UGA) and ochre (UAA), due to an alteration in its anticodon,15, 20 these agents have high stop codon specificity and also show higher nonsense suppression efficiency than other readthrough strategies.21 Thus, sup-tRNAs emerge as interesting candidates to be used as therapeutic agents to recover the expression of full-length functional proteins. Several studies demonstrated efficient nonsense suppression in mammalian models, although the recovery of functional proteins was rarely shown.15, 20, 22, 23, 24, 25
In this work we wish to: (1) evaluate the potential impact of using nonsense suppression in HDGC; (2) determine the number of tRNAs necessary to treat nonsense mutation-carrying HDGC families; and (3) assess the possibility of recovering expression of a functional protein from an allele carrying a nonsense mutation described in HDGC using a sup-tRNA. To our knowledge, this is the first demonstration of functional recovery of a causative gene in cancer by cognate amino acid replacement with sup-tRNAs.
Materials and methods
Plasmid construction
CDH1 mini-gene
WTcDNA pIRES-EGFP2 vector, previously designed in our lab,26 was used to obtain a CDH1 WT mini-gene (WTint), enclosing an intron 15 minimal sequence (291 bp) between CDH1 exons 15 (L34788.1) and 16 (L34789.1).
Modified pIRES vector with sup-tRNAArg
A new multiple cloning site (MCS) was inserted into pIRES-EGFP2 vector, 919 bp upstream the CMV promoter (Figure 1a and Supplementary Figure S1). Arginine–tRNA was then inserted into this vector and mutated by SDM. Detailed procedures and list of primers can be found in Supplementary Data and Supplementary Table S1.
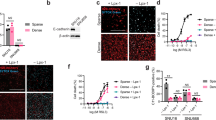
Suppressor-tRNA restores E-cadherin expression in AGS cell line. (a) Schematic representation of the plasmids used: pIRES-EGFP2 vector with a MCS distant from CMV promoter to allow for stable tRNA expression (left side); and from a vector with the sequence of both Sup-tRNAArg gene and CDH1 mini-gene (right side). (b) The tRNAUCGArg secondary structure indicating the mutation introduced in the anticodon (left side); the mutant tRNAUCGArg (Sup-tRNAArg) is able to decode the PTC as arginine originating a full-length protein (right side). (c, d) AGS cells transiently transfected with pIRES-EGFP2-modified vectors: mock-MCS, 1003int, 1003int_sup-tRNAArg, Sup-tRNAArg, WTint, WTint_sup-tRNAArg, 1003_cDNA, 1003_cDNA_sup-tRNAArg, WTcDNA, WTcDNA_sup-tRNAArg and parental cells, treated and not treated with Lipofectamine 2000. (c) RNA expression was analysed by RT-PCR for CDH1 transcripts, using GFP expression for transfection control, and GAPDH as endogenous RNA control; protein expression was detected by WB using as endogenous control α-tubulin. (d) Immunocytochemistry in cells transfected with 1003int_sup-tRNAArg and 1003_cDNA_sup-tRNAArg show E-cadherin expression localized at the membrane, whereas control cells 1003int and 1003_cDNA do not have any expression.
Cell culture
AGS, MDA-MB-231 and CHO cells were cultivated in RPMI, DMEM and α-MEM media supplemented with 1% penicillin–streptomycin (Gibco, Green Island, NY, USA), respectively, and 10% FBS (Lonza, Walkersville, MD, USA) at 37 °C with 5% CO2.
Transfections
Transient and stable transfections were carried using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Stably expressing cells were selected in 1 mg/ml Geneticin (Gibco) 72 h after transfection.
Nucleic acid extraction and cDNA synthesis
DNA was extracted with Invisorb Spin Tissue Mini Kit (Invitek, Berlin, Germany); RNA was isolated with Tripure Isolation Reagent (Roche, Mannheim, Germany); and first-strand cDNA was synthesized from 1 μg of total RNA with Superscript II Reverse Transcriptase and random hexamer primers (Invitrogen), all according to the manufacturer’s instructions.
Protein extraction and western blotting
Western blots were performed, using total protein lysates, with antibodies against E-cadherin (1:1000; SHE78-7, Enzo Life Sciences Inc., Farmingdale, NY, USA), GFP (1:500, Santa Cruz Biotechnology Inc., Dallas, TX, USA) and α-tubulin (1:10 000, Sigma, St Louis, MO, USA), as described in Supplementary Data.
Immunocytochemistry
E-cadherin expression was analysed with SHE78-7 (1:100, Enzo Life Sciences Inc.) in fixed cells (4% formaldehyde), as described in Supplementary Data.
Cell sorting and flow cytometry
E-cadherin-expressing cells were analysed by flow cytometry using anti-E-cadherin HECD1 (1:100; Zymed Laboratories Inc., Camarillo, CA, USA) and anti-mouse Alexa Fluor 647 (1:500; Invitrogen). Detailed protocol is in Supplementary Data.
Proximity ligation assay (PLA)
This assay was performed as described previously.27
Statistical analysis
Two-tailed paired Student’s t-test from GraphPad Prism 5 (San Diego, CA, USA) was used. In all analyses, P<0.05 was required for statistical significance.
Results
One-fifth of all CDH1 germline mutation carriers and their families may benefit from nonsense suppression therapy
We revised all deleterious CDH1 germline mutations so far described in the literature in E-cadherin-related diseases. These mutations have been described in 108 HDGC and 5 LBC families, and in 10 patients with EODGC (Table 1). Nonsense mutations occurred in 20% (n=25) of these cases (23 HDGC, 1 LBC and 1 EODGC).4, 28, 29, 30 Under the hypothesis raised in the present work, all related nonsense mutation carriers may benefit from a target-designed sup-tRNA strategy (Table 1).
The 25 cases carrying nonsense CDH1 mutations carried 17 different mutations that converted seven different amino acids into stop codons: arginine (Arg), glutamine (Gln), glycine (Gly), glutamic acid (Glu), tryptophan (Trp), tyrosine (Tyr) and cysteine (Cys) (Table 2). Arginine, the most frequently affected amino acid, was mutated to a stop codon in nine families, and therefore could be treated with a single sup-tRNAArg (Table 3). Glutamine, the second most frequently affected amino acid that is replaced by an amber stop codon, could be replaced by a sup-tRNAGln in seven cases. An opal sup-tRNAGln would be necessary to treat 1 family. Tryptophan, glutamic acid and glycine sup-tRNAs can suppress PTCs in two families each. Tyrosine and cysteine sup-tRNAs could be used to treat two cases (Table 3). In summary, a tRNA-based strategy would be useful to treat ∼20% of HDGC, LBC families and EODGC cases, using only 8 different sup-tRNAs (Table 3).
Development of a model system to simultaneously express mutant CDH1 and a sup-tRNA
As arginine was the amino acid that most frequently got mutated to a stop codon in HDGC families, we chose one HDGC-related nonsense mutation that replaced an Arg by a PTC at position 1003 of the CDH1 coding sequence (c.1003C>T) in exon 7 (L34793.1) of the CDH1 gene31 for studying tRNA nonsense suppression. We used the pIRES-EGFP2 backbone carrying either the CDH1 cDNA sequence (WTcDNA)26 or a CDH1 mini-gene (WTint) in which a minimal intronic sequence from canonical CDH1 intron 15 was introduced (R Bordeira-Carriço et al, unpublished). The inclusion of an intronic sequence in this construct generated an artificial CDH1 pre-mRNA that underwent splicing and endogenous quality control (Figure 1c). Both WTcDNA and WTint sequences were mutagenized by SDM, generating the 1003cDNA and 1003int mutant mini-gene-carrying vectors. These vectors were further engineered to simultaneously express CDH1 sequences (WT and mutant) and the sup-tRNAArg (sup-tRNAArg). Several critical features of tRNA genes biology were considered during the engineering of this plasmid, namely: (1) tRNA gene transcription by RNA polymerase III (Pol III), and not Pol II that transcribes our CDH1 gene; (2) tRNA internal promoter elements located within the transcribed region; and (3) upstream sequence motifs at −50 to −60 bp and TATA box at −30 bp relative to transcription start site that modulate tRNA transcription.32 To match these requirements, we designed (Supplementary Figure S1) and inserted a new MCS upstream of the CMV promoter in which the Arg tRNA gene sequence was cloned (Figure 1a). The anticodon (UCG) of the Arg tRNA gene was mutated to UCA by SDM, originating a supressor-tRNAUCG→UCAArg (sup-tRNAArg) that could decode the 1003C>T nonsense mutation by standard Watson–Crick decoding (Figure 1b). This modified pIRES-EGFP2 vector, carrying both the sup-tRNAArg and mutant or WT CDH1 sequences, was designed to ensure that all cells would express both genes. Variants of this vector were also produced containing five copies of the sup-tRNAArg gene (sup-tRNA5Arg), associated or not with the 1003cDNA and the WTcDNA, to analyse the effect of sup-tRNAArg copy number in E-cadherin expression recovery in a controlled system. The sup-tRNAArg was expected to promote the insertion of Arg at the PTC site, leading to the production of WT E-cadherin (Figure 1b).
Induced expression of sup-tRNAArg led to recovery of E-cadherin expression
All control vectors produced were transfected into AGS cell line to generate the following cell lines: Mock (empty vector), WTcDNA, WTint, sup-tRNA, 1003cDNA, 1003int, WTcDNA_sup-tRNA, WTint_sup-tRNA and WTcDNA_sup-tRNA5Arg. Expression of both GFP and CDH1 genes was determined in transiently transfected AGS cells (Figure 1c) and two other cell lines (MDA-MB-231 and CHO, Supplementary Figure S2). CDH1 was residually expressed in parental AGS cells as well as in Mock or sup-tRNA-expressing cells (Figure 1c). In contrast, the transfection of any vector containing CDH1 sequences led to an increase in the CDH1 mRNA expression. GFP expression was detected in all cell lines transfected with pIRES-EGFP2 vector, but not in nontransfected controls (parental and lipofectamine; Figure 1c).
Importantly, in terms of protein expression we showed that: (1) positive controls expressed high levels of correctly processed E-cadherin (WTcDNA, WTint); (2) none of the 1003 mutant vectors expressed E-cadherin (1003cDNA, 1003int); (3) the expression of sup-tRNA alone did not induce E-cadherin expression (sup-tRNA); and (4) one or more copies of sup-tRNA did not modify WT E-cadherin expression levels or processing (WTcDNA_sup-tRNA, WTint_sup-tRNA and WTcDNA_sup-tRNA5Arg; Figure 1c).
We next assessed the in vitro E-cadherin expression recovery from a CDH1 PTC-carrying mRNA by the sup-tRNAArg. The expression of sup-tRNAArg was able to restore E-cadherin protein expression in AGS cells carrying either the mutant mini-gene (1003int_sup-tRNAArg) or the mutant CDH1 cDNA sequence (1003_cDNA_sup-tRNAArg; Figure 1c). This experiment was reproduced several times with the same results in AGS cells, as well as in MDA-MB-231 and CHO cells expressing the 1003int_sup-tRNAArg vector (Supplementary Figure S2). In all cell lines expressing the sup-tRNAArg, the molecular weight of E-cadherin molecules, after sup-tRNAArg correction, was similar to that obtained for control WTint and WTcDNA cells (Figure 1c and Supplementary Figure S2). This result shows that sup-tRNAArg was able to correct E-cadherin mutant sequences generating full-length and correctly processed E-cadherin molecules, as demonstrated by the observation of a high molecular weight band in the western blot (representing the mature form of E-cadherin; Figure 1c and Supplementary Figure S2).
Protein expression levels obtained after tRNA suppression were low when compared with WT controls (Figure 1c). Nevertheless, the recovered E-cadherin protein was correctly localized at the cell membrane in AGS, CHO and MDA-MB-231 cell lines and presented an expression pattern similar to that observed in E-cadherin WT-expressing cells, as observed by immunocytochemistry (Figure 1d and Supplementary Figure S2). Moreover, flow cytometry clearly confirmed the correct E-cadherin membranous localization (Figure 2a). Flow cytometry analysis further revealed that sup-tRNAArg expression from the single tRNA gene copy construct was sufficient to recover membranous E-cadherin expression in ∼30% of AGS cells expressing either the mutant mini-gene (1003int_sup-tRNAArg) or the mutant cDNA (1003cDNA_sup-tRNAArg), in comparison with WTint or WTcDNA vectors (Figure 2a). The sup-tRNAArg induced a twofold increase in 1003int and 1003cDNA cells (Figure 2b). A vector carrying the 1003cDNA and five copies of sup-tRNAArg (1003cDNA_sup-tRNA5Arg) transfected into AGS cells induced a slight increase in the percentage of E-cadherin-positive cells and in the intensity of E-cadherin membranous expression, although this was not statistically significant (Figures 2a and b). In summary, the tested sup-tRNAArg was able to restore E-cadherin protein expression, at the correct cellular location, to levels similar to those displayed by E-cadherin WT-expressing cells, at least in 30% of the transfected cells.
Flow cytometry for transfected AGS cells. This analysis shows membranous E-cadherin expression in 1003int_sup-tRNAArg, WTint, 1003_cDNA_sup-tRNAArg, 1003_sup-tRNA5Arg and WTcDNA. (a) Graph bars for CDH1 mini-gene (left) and CDH1 cDNA (right) vectors are shown as percentage of membranous E-cadherin expressing cells (values normalized for Mock). (b) E-cadherin expression intensity in the analysed population, using geometric mean of population intensity parameter, was also analysed for CDH1 mini-gene (left) and CDH1 cDNA (right) vectors. The graphs show the mean±SD of at least four independent experiments. Statistical significance is depicted in graphs for P≤0.001 (***).
Induced expression of sup-tRNAArg led to recovery of adherens junction complex (AJC) formation
To assess whether sup-tRNAArg-recovered E-cadherin membranous expression from mutant plasmids could correctly establish adherens junctions (AJ) through the formation of complexes with β-catenin and p120-catenin, we used the PLA. This technique has been successfully used by our group to distinguish pathogenic missense E-cadherin mutations from nonpathogenic or WT E-cadherin.27 Following the same protocol, a striking increase was seen in the number of E-cadherin interactions with β-catenin and p120-catenin when sup-tRNAArg was coexpressed with 1003int and 1003cDNA (Figure 3) in comparison with that of cells expressing the same vectors lacking sup-tRNAArg. These interaction levels were yet lower than those observed for E-cadherin WT-expressing cells. Most likely, cells undergoing sup-tRNAArg correction become scattered among noncorrected/nontransfected cells, diminishing the probability of AJC formation with adjacent E-cadherin-expressing cells.
E-cadherin restored by sup-tRNAArg recruits the adhesion-complex molecules β-catenin and p120-catenin. The interaction between E-cadherin and β-catenin (upper panel) or p120-catenin (lower panel) was assessed by in situ PLA in AGS cells. Positive signals were detected in 1003int_sup-tRNAArg, WTint, 1003_cDNA_sup-tRNAArg and WTcDNA cells (amplification × 40).
Taken together, these results demonstrate that sup-tRNAArg is able to induce the production of full-length E-cadherin from a mutant gene harbouring a PTC, and that PTC readthrough leads to correct recruitment of β-catenin and p120-catenin AJC components.
Discussion
HDGC is an aggressive disease, difficult to diagnose and with a high risk for GC development in CDH1 germline mutation carriers.1, 9 As in many other cancer syndromes, very few therapeutic options beyond surgery and conventional chemo- and radio-therapy exist, and the success of these conventional approaches is limited.33, 34, 35 Many CDH1 mutation carriers refuse prophylactic gastrectomy, and no alternatives exist to protect these individuals from developing cancer. We hypothesize that nonsense suppression could constitute an alternative for these patients by promoting the recovery of full-length E-cadherin expression. Recent studies have shown the recovery of P53 expression36 and APC37 expression from a nonsense mutant by aminoglycosides and a macrolide antibiotic in cancer cell lines and in a colorectal cancer mouse model, with reduction of the oncogenic phenotype.37 Although promising, these strategies present a great conceptual problem as the use of unspecific readthrough strategies leads to replacement of a nonsense codon by a near-cognate amino acid. In practical terms, this represents the introduction of a missense mutation that may be deleterious in most cancer-associated syndromes.10 The use of sup-tRNAs allows circumventing this problem as tRNA-mediated nonsense suppression can lead to incorporation of the cognate, and therefore WT, amino acid at the PTC site.10
To analyse the frequency of nonsense mutations in HDGC spectrum, we performed an extensive analysis of CDH1 mutations described to date in HDGC, LBC and EODGC, and identified 25 families with nonsense mutations. Only eight different sup-tRNAs would be required to induce nonsense suppression in these patients and their families. As arginine was the most frequently affected amino acid, we selected an arginine-sup-tRNA to study the possibility of recovering E-cadherin expression from a mutant CDH1 gene in different cell line models.
Using CDH1 WT and mutant in vitro models, we demonstrated that sup-tRNAArg restored the production of E-cadherin protein expression in cancer and noncancer cell lines. This full-length, putatively functional E-cadherin was correctly localized at the cell membrane and was able to promote AJC formation via the binding to β-catenin and p120-catenin.
Suppression of nonsense mutations by sup-tRNAs has been demonstrated in Xenopus oocytes38 and in mammalian cells,20, 22 and was studied later as putative therapeutic tools for diseases like β-thalassaemia,23 xeroderma pigmentosum24 and DMD.15 Nevertheless, formal proof of functional recovery was rarely reported. The closest demonstration of function recovery was described for chloramphenicol acetyltransferase (CAT) carrying a nonsense mutation, whose expression by a multimerized sup-tRNA was observed in a transgenic mouse model.25 The present study constitutes the first report of efficient nonsense suppression induced by a sup-tRNA in a human cancer-associated gene, with recovery of full-length and likely functional protein. Importantly, we have shown that ∼30% of sup-tRNAArg-expressing cells recovered E-cadherin membranous expression. This is similar to results obtained with aminoglycosides and PTC124 (drugs tested in clinical trials) in cultured cell lines and mouse models of several diseases, where the efficiency of production of full-length functional protein ranged from 1 to 25%.15, 16 Reasons for this low efficiency are the dependence on PTC identity,39 PTC context,40 activity of translation release factor 1 (eRF1)21 and the decoding efficiency of the sup-tRNA.40 A question raised upon use of readthrough strategies is its effect on global translation efficiency, namely the possibility of suppressing canonical stop codons. It seems that suppression agents are more effective at PTCs than at canonical stop codons.41 Translation termination at PTCs and canonical stop codons may differ (1) because of longer ribosomal pause at PTCs and (2) also because canonical stop codons and surrounding contexts are less susceptible to suppression.41 In addition, in-frame stop codons coexist at the end of many ORFs, reducing the possibility of translation elongation into the 3′ UTR.42 These arguments support the idea of higher suppression efficiency at PTCs.41 Moreover, mRNA turnover pathways, such as the Nonstop mRNA decay, often target mRNAs with long 3′ UTRs for degradation.43 These observations suggest that a nonsense suppression strategy could be applied with a reduced toxic effect resulting from readthrough of canonical stop codons.
To our knowledge, this is the first report of the use of a sup-tRNA to restore full-length protein expression and function of a human cancer-associated gene. Furthermore, it supports the possibility of using this strategy for cancer prevention. In germline mutation carriers, sup-tRNAs could allow WT protein expression from the mutant allele, reducing the risk of cancer development, even if a second somatic event inactivates the remaining WT allele. Importantly, the 20% of HDGC families carrying CDH1 nonsense mutations enclose many mutation carriers that are potential candidates for correction with the sup-tRNAArg. Furthermore, this strategy could be applied to other cancer syndromes beyond HDGC in which nonsense mutations constitute from 10 to 30% of all mutations described (recently reviewed in Bordeira-Carrico et al10). Although each of these syndromes have a very low incidence within the population, when combined, they comprise a significant portion of germline mutation carriers to which an alternative and nonsurgical approach could be offered. Moreover, sup-tRNAs could constitute a therapeutic tool for diseases caused by nonsense mutations, beyond Duchenne muscular dystrophy and cystic fibrosis, such as β-thalassaemia,44 neurodegenerative diseases45 or haemophilia, where the efficiency of aminoglycosides or PTC124 is yet to be demonstrated.10
The use of sup-tRNA-based therapy in clinical practice faces many challenges, especially regarding the efficiency of delivery and expression of these systems in vivo, with a minimal toxicity caused to the organism. Plasmid-based systems and viral vectors raise several issues, such as toxicity, the risk of insertional mutagenesis, gene silencing or the induction of immune response within the organism.46 A possible alternative that might overcome these problems are the recently developed minicircles, supercoiled DNA molecules free of bacterial plasmid backbone elements.47 Additionally, an interesting hypothesis for the delivery of these vectors in vivo would be the use of nanoparticles, a field with promising advances in the past years. Nonviral vectors based on liposomes and polymers of different materials, as for instance PEI48 or chitosan,49, 50 allow the delivery of nucleotides with greater flexibility than viral vectors, in terms of the size of the nucleotide cargo that can be transported.48 In this way, they can bypass the immune system with fewer safety concerns, may allow targeted delivery through receptor-mediated uptake into the cell of interest48 and can be orally available.49 A sup-tRNA encoding gene is small and can be easily integrated into a vector, and can use the endogenous machinery to produce the tRNAs, as we demonstrated in this work. It would be possible to complex a sup-tRNA gene into a nanoparticle to improve the delivery efficiency in vivo. Further developments in nanomedicine will most certainly turn the use of sup-tRNAs as a therapeutic tool in clinical practice into reality.
References
Fitzgerald RC, Hardwick R, Huntsman D et al: Hereditary diffuse gastric cancer: updated consensus guidelines for clinical management and directions for future research. J Med Genet 2010; 47: 436–444.
Oliveira C, Pinheiro H, Figueiredo J, Seruca R, Carneiro F : E-cadherin alterations in hereditary disorders with emphasis on hereditary diffuse gastric cancer. Prog Mol Biol Transl Sci 2013; 116: 337–359.
Guilford P, Hopkins J, Harraway J et al: E-cadherin germline mutations in familial gastric cancer. Nature 1998; 392: 402–405.
Oliveira C, Senz J, Kaurah P et al: Germline CDH1 deletions in hereditary diffuse gastric cancer families. Hum Mol Genet 2009; 18: 1545–1555.
Masciari S, Larsson N, Senz J et al: Germline E-cadherin mutations in familial lobular breast cancer. J Med Genet 2007; 44: 726–731.
Corso G, Pedrazzani C, Pinheiro H et al: E-cadherin genetic screening and clinico-pathologic characteristics of early onset gastric cancer. Eur J Cancer 2011; 47: 631–639.
Kaurah P, MacMillan A, Boyd N et al: Founder and recurrent CDH1 mutations in families with hereditary diffuse gastric cancer. JAMA 2007; 297: 2360–2372.
Oliveira C, Seruca R, Carneiro F : Genetics, pathology, and clinics of familial gastric cancer. Int J Surg Pathol 2006; 14: 21–33.
Oliveira C, Seruca R, Carneiro F : Hereditary gastric cancer. Best Pract Res Clin Gastroenterol 2009; 23: 147–157.
Bordeira-Carrico R, Pego AP, Santos MS, Oliveira C : Cancer syndromes and therapy by stop-codon readthrough. Trends Mol Med 2012; 18: 667–678.
Carneiro F, Huntsman DG, Smyrk TC et al: Model of the early development of diffuse gastric cancer in E-cadherin mutation carriers and its implications for patient screening. J Pathol 2004; 203: 681–687.
Oliveira C, Sousa S, Pinheiro H et al: Quantification of epigenetic and genetic 2nd hits in CDH1 during hereditary diffuse gastric cancer syndrome progression. Gastroenterology 2009; 136: 2137–2148.
Huntsman DG, Carneiro F, Lewis FR : Early gastric cancer in young, asymptomatic carriers of germ-line E-cadherin mutations. N Engl J Med 2001; 344: 1904–1909.
Oliveira C, Seruca R, Hoogerbrugge N, Ligtenberg M, Carneiro F : Clinical utility gene card for: hereditary diffuse gastric cancer (HDGC). Eur J Hum Genet 2013; 21: doi:10.1038/ejhg.2012.247.
Kellermayer R : Translational readthrough induction of pathogenic nonsense mutations. Eur J Med Genet 2006; 49: 445–450.
Welch EM, Barton ER, Zhuo J et al: PTC124 targets genetic disorders caused by nonsense mutations. Nature 2007; 447: 87–91.
Figueiredo J, Simoes-Correia J, Soderberg O, Suriano G, Seruca R : ADP-ribosylation factor 6 mediates E-cadherin recovery by chemical chaperones. PLoS One 2011; 6: e23188.
Mateus AR, Simoes-Correia J, Figueiredo J et al: E-cadherin mutations and cell motility: a genotype-phenotype correlation. Exp Cell Res 2009; 315: 1393–1402.
Simoes-Correia J, Figueiredo J, Oliveira C et al: Endoplasmic reticulum quality control: a new mechanism of E-cadherin regulation and its implication in cancer. Hum Mol Genet 2008; 17: 3566–3576.
Capone JP, Sedivy JM, Sharp PA, RajBhandary UL : Introduction of UAG, UAA, and UGA nonsense mutations at a specific site in the Escherichia coli chloramphenicol acetyltransferase gene: use in measurement of amber, ochre, and opal suppression in mammalian cells. Mol Cell Biol 1986; 6: 3059–3067.
Diop D, Chauvin C, Jean-Jean O : Aminoglycosides and other factors promoting stop codon readthrough in human cells. C R Biol 2007; 330: 71–79.
Sako Y, Usuki F, Suga H : A novel therapeutic approach for genetic diseases by introduction of suppressor tRNA. Nucleic Acids Symp Ser (Oxf) 2006; 50: 239–240.
Temple GF, Dozy AM, Roy KL, Kan YW : Construction of a functional human suppressor tRNA gene: an approach to gene therapy for beta-thalassaemia. Nature 1982; 296: 537–540.
Panchal RG, Wang S, McDermott J, Link CJ Jr : Partial functional correction of xeroderma pigmentosum group a cells by suppressor tRNA. Hum Gene Ther 1999; 10: 2209–2219.
Buvoli M, Buvoli A, Leinwand LA : Suppression of nonsense mutations in cell culture and mice by multimerized suppressor tRNA genes. Mol Cell Biol 2000; 20: 3116–3124.
Pinheiro H, Carvalho J, Oliveira P et al: Transcription initiation arising from E-cadherin/Cdh1 Intron2: a novel protein isoform that increases gastric cancer cell invasion and angiogenesis. Hum Mol Genet 2012; 21: 4253–4269.
Figueiredo J, Soderberg O, Simoes-Correia J, Grannas K, Suriano G, Seruca R : The importance of E-cadherin binding partners to evaluate the pathogenicity of E-cadherin missense mutations associated to HDGC. Eur J Hum Genet 2012; 21: 301–309.
Pinheiro H, Bordeira-Carrico R, Seixas S et al: Allele-specific CDH1 downregulation and hereditary diffuse gastric cancer. Hum Mol Genet 2010; 19: 943–952.
Guilford P, Humar B, Blair V : Hereditary diffuse gastric cancer: translation of CDH1 germline mutations into clinical practice. Gastric Cancer 2010; 13: 1–10.
Carvalho J, Pinheiro H, Oliveira C : E-cadherin germline mutations; in Corso G, Roviello F (eds): Spotlight on Familial and Hereditary Gastric Cancer. New York: Springer, 2013, pp 35–49.
Jonsson BA, Bergh A, Stattin P, Emmanuelsson M, Gronberg H : Germline mutations in E-cadherin do not explain association of hereditary prostate cancer, gastric cancer and breast cancer. Int J Cancer 2002; 98: 838–843.
Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A : The expanding RNA polymerase III transcriptome. Trends Genet 2007; 23: 614–622.
Cervantes A, Roda D, Tarazona N, Rosello S, Perez-Fidalgo JA : Current questions for the treatment of advanced gastric cancer. Cancer Treat Rev 2013; 39: 60–67.
Dicken BJ, Bigam DL, Cass C, Mackey JR, Joy AA, Hamilton SM : Gastric adenocarcinoma: review and considerations for future directions. Ann Surg 2005; 241: 27–39.
Finch A, Evans G, Narod SA : BRCA carriers, prophylactic salpingo-oophorectomy and menopause: clinical management considerations and recommendations. Womens Health (Lond Engl) 2012; 8: 543–555.
Floquet C, Deforges J, Rousset JP, Bidou L : Rescue of non-sense mutated p53 tumor suppressor gene by aminoglycosides. Nucleic Acids Res 2011; 39: 3350–3362.
Zilberberg A, Lahav L, Rosin-Arbesfeld R : Restoration of APC gene function in colorectal cancer cells by aminoglycoside- and macrolide-induced read-through of premature termination codons. Gut 2010; 59: 496–507.
Rodriguez EA, Lester HA, Dougherty DA : In vivo incorporation of multiple unnatural amino acids through nonsense and frameshift suppression. Proc Natl Acad Sci USA 2006; 103: 8650–8655.
Linde L, Kerem B : Introducing sense into nonsense in treatments of human genetic diseases. Trends Genet 2008; 24: 552–563.
Phillips-Jones MK, Hill LS, Atkinson J, Martin R : Context effects on misreading and suppression at UAG codons in human cells. Mol Cell Biol 1995; 15: 6593–6600.
Keeling KM, Bedwell DM : Suppression of nonsense mutations as a therapeutic approach to treat genetic diseases. Wiley Interdiscip Rev RNA 2011; 2: 837–852.
Dalphin ME, Stockwell PA, Tate WP, Brown CM : Transterm, the translational signal database, extended to include full coding sequences and untranslated regions. Nucleic Acids Res 1999; 27: 293–294.
Frischmeyer PA, van Hoof A, O'Donnell K, Guerrerio AL, Parker R, Dietz HC : An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 2002; 295: 2258–2261.
Cao A, Galanello R : Beta-thalassemia. Genet Med 2010; 12: 61–76.
Mitui M, Bernatowska E, Pietrucha B et al: ATM gene founder haplotypes and associated mutations in Polish families with ataxia-telangiectasia. Ann Hum Genet 2005; 69: 657–664.
Jechlinger W : Optimization and delivery of plasmid DNA for vaccination. Expert Rev Vaccines 2006; 5: 803–825.
Osborn MJ, McElmurry RT, Lees CJ et al: Minicircle DNA-based gene therapy coupled with immune modulation permits long-term expression of alpha-L-iduronidase in mice with mucopolysaccharidosis type I. Mol Ther 2011; 19: 450–460.
Oliveira H, Fernandez R, Pires LR et al: Targeted gene delivery into peripheral sensorial neurons mediated by self-assembled vectors composed of poly(ethylene imine) and tetanus toxin fragment c. J Control Release 2010; 143: 350–358.
Oliveira CR, Rezende CM, Silva MR, Borges OM, Pego AP, Goes AM : Oral vaccination based on DNA-chitosan nanoparticles against Schistosoma mansoni infection. ScientificWorldJournal 2012; 2012: 938457.
Pires LR, Oliveira H, Barrias CC et al: Imidazole-grafted chitosan-mediated gene delivery: in vitro study on transfection, intracellular trafficking and degradation. Nanomedicine (Lond) 2011; 6: 1499–1512.
Acknowledgements
This work has been supported by: the Calouste Gulbenkian Foundation (Project: ‘Mutated suppressor tRNAs as a therapeutic tool for cancer associated syndromes: HDGC as a model’); COMPETE/FEDER FCT (project: PTDC/SAU-GMG/098850/2008); FCT PhD grant SFRH/BD/46462/2008 to RB-C, research grant from project PTDC/SAU-GMG/72168/2006 to DF, postdoctoral grant from project FCT PTDC/SAU-GMG/110785/2009 to DDM and FCT postdoctoral grant to HP. We acknowledge Ana Sofia Ribeiro, Joana Figueiredo, Marta L Pinto and Patrícia Pereira for technical support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Bordeira-Carriço, R., Ferreira, D., Mateus, D. et al. Rescue of wild-type E-cadherin expression from nonsense-mutated cancer cells by a suppressor-tRNA. Eur J Hum Genet 22, 1085–1092 (2014). https://doi.org/10.1038/ejhg.2013.292
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2013.292
Keywords
This article is cited by
-
Engineered tRNAs suppress nonsense mutations in cells and in vivo
Nature (2023)
-
Repurposing tRNAs for nonsense suppression
Nature Communications (2021)
-
Nonsense suppression therapies in human genetic diseases
Cellular and Molecular Life Sciences (2021)
-
Advances in therapeutic use of a drug-stimulated translational readthrough of premature termination codons
Molecular Medicine (2018)