Abstract
AZF microdeletion screening is routinely performed in the diagnostic work-up for male infertility; however, some issues remain debated. In this study, we provide insights into the sperm concentration cutoff value for routine testing, the predictive value of AZFc deletion for testicular sperm retrieval and the Y-background contribution to the interpopulation variability of deletion frequencies. In the Spanish population, partial AZFc rearrangements have been poorly explored and no data exist on partial duplications. In our study, 27/806 (3.3%) patients carried complete AZF deletions. All were azoo/cryptozoospermic, except for one whose sperm concentration was 2 × 106/ml. In AZFc-deleted men, we observed a lower sperm recovery rate upon conventional TESE (9.1%) compared with the literature (60–80% with microTESE). Haplogroup E was the most represented among non-Spanish and hgr P among Spanish AZF deletion carriers. The analysis of AZFc partial rearrangements included 330 idiopathic infertile patients and 385 controls of Spanish origin. Gr/gr deletion, but not AZFc partial duplications, was significantly associated with spermatogenic impairment. Our data integrated with the literature suggest that: (1) routine AZF microdeletion testing could eventually include only men with ≤2 × 106/ml; (2) classical TESE is associated with low sperm recovery rate in azoospermic AZFc-deleted men, and therefore microTESE should be preferred; (3) Y background could partially explain the differences in deletion frequencies among populations. Finally, our data on gr/gr deletion further support the inclusion of this genetic test in the work-up of infertile men, whereas partial AZFc duplications do not represent a risk for spermatogenic failure in the Spanish population.
Similar content being viewed by others
Introduction
Y-chromosome microdeletions are a well-established genetic cause of severe spermatogenic failure and their molecular diagnosis is part of the diagnostic work-up of severe male factor infertility.1 These submicroscopic deletions, involving the AZF region of the Yq, can be classified according to the recombination hot spot and have been designated as AZFa, P5-proximal P1 (AZFb), P5-distal P1 (AZFbc), P4-distal P1 (AZFbc) and b2/b4 (AZFc).2, 3 The deletion frequency clearly varies according to the semen phenotype; indeed, severely oligozoospermic and azoospermic men have the highest risk of carrying Y microdeletions. The different deletion frequency observed even within similar semen categories among infertile men from different populations suggests that ethnic background could also influence the occurrence of this genetic anomaly. The lowest deletion frequency (1.8%) was reported in German and Danish idiopathic severely oligozoospermic men,1, 4 whereas the highest was reported in an ethnically admixed population from France (13.7%)5 and in Romanians (10%).6 Data on the prevalence of classical AZF deletions in men attending an infertility clinic in Spain derive from two independent surveys, with an overall frequency of 5.4% and 7%, respectively.7, 8
Because of its complex structure, rich in massive near-identical amplicons, the AZFc region is particularly susceptible to homology-based intrachromosomal recombination events and hence to structural variations as copy number variations (CNVs).9, 10 In addition to the classical AZFc deletion, several recurrent partial deletions (named gr/gr, b2/b3 and b1/b3) and duplications (b2/b4 duplication) have been reported.11, 12 Even though all partial rearrangements produce either a decrease or an increase in AZFc gene dosage, only the ‘gr/gr deletion’ resulted to be clinically relevant. The clinical significance of this recurrent deletion has been object of a long-lasting debate. Controversies are mainly because of selection biases and the lack of ethnic matching between cases and controls.13 Notwithstanding, the four meta-analyses published so far on this topic indicate that gr/gr deletion represents a significant risk factor for impaired sperm production.14, 15, 16, 17 The clinical relevance of gr/gr deletion has been confirmed further by a recent multiethnic population-based survey of >20 000 Y chromosomes, reporting a 1.9-fold increased risk of severe spermatogenic failure.18 The prevalence and clinical significance of partial AZFc rearrangements in the Spanish population has been little explored and only one pilot study was performed.19 Similarly, partial AZFc duplications in male infertility have been poorly explored. To date, only two groups have addressed this topic in the Taiwanese and Italian populations, reaching contradictory conclusions.12, 20 In addition, by using a consecutive cohort study model, Noordam et al21 suggested that both lower and higher DAZ gene dosage could be deleterious for spermatogenesis.
This study presents the 8-year experience of our clinic in testing infertile men for Y-linked CNVs. Our first aim was to thoroughly describe the genetic makeup (karyotype and classical AZF deletions) of consecutive infertile men referring to our genetic laboratory and, thus, to provide further data on the clinical indications for routine genetic testing. Our second aim was to corroborate the clinical relevance of gr/gr deletion in Spain by performing a detailed molecular characterization of the AZFc region in a carefully matched case/control study setting.
For the first time, we provide data on the prevalence and clinical significance of AZFc partial duplications in the Spanish population, contributing to outline the effect of an increased DAZ gene dosage on sperm production in a Caucasian Y background.
Patients and methods
Subjects
We retrospectively analyzed a set of 806 consecutive infertile men, screened for Y-chromosome microdeletions between November 2004 and December 2012. Most of them (72.95%) were Spanish, whereas the remaining (27.05%) were of different geographic origin. The majority of non-Spanish patients (53.7%) came from North-Western Africa, mostly from Morocco (44.0%). The second most represented countries were Southern and Central America (22%), followed by Middle and Far East (9.2%), with Eastern, North-Western and Southern Europe accounting for 7.3%, 6.0% and 1.8% of non-Spanish patients, respectively. All patients underwent a comprehensive andrological examination (including physical examination, scrotal ultrasound and hormone analysis) and karyotype analysis was performed for 747 men. Based on clinical and karyotype data, patients were classified into ‘idiopathic’ and ‘nonidiopathic’ (Table 1), except for 27 (3.3%) patients whose medical history resulted insufficient for an etiologic classification. Semen analysis was performed according to the WHO guidelines22 except for morphology, for which strict criteria were used. In all, 291 patients were azoospermic (AZ); 392 and 88 presented severe (SOZ; 0<sperm concentration (SC) ≤5 × 106/ml) and moderate oligozoospermia (MOZ; 5<SC<20 × 106/ml), respectively, 31 had normal SC (≥20 × 106/ml) but low motility (asthenozoospermia) or <4% of normal morphology (teratozoospermia) or a combination of both (asthenoteratozoospermia). For four patients, semen parameters were not available. Bilateral testicular biopsy was performed in 213 patients. A single biopsy was retrieved after scrotal incision from each testis for both diagnostic (to define the type of tubular damage) and therapeutic purposes (to recover spermatozoa for assisted reproductive techniques (ARTs)). Overall, mature sperm could be retrieved in 45.1% of cases (for further details, see Supplementary Table 1).
Study population for the screening of partial AZFc rearrangements
From a total of 715 Spanish subjects, 330 strictly selected ‘idiopathic’ infertile patients and 385 controls were analyzed for partial AZFc rearrangements. This group included: 94 AZ, 190 SOZ and 46 MOZ men. Controls were recruited on the basis of normal sperm parameters22 among sperm donors and men with proven fertility. The total motile sperm count (TMC) was calculated for all subjects by multiplying semen volume by sperm concentration and the percentage of progressively motile spermatozoa.
To prevent recruitment bias, much care was taken for the ethnic and geographic matching of patients and controls. All were explicitly asked for their paternal and maternal origin and only subjects with proven Spanish ancestry were included. The Y-chromosome haplogroup (hgr) analysis further confirmed the similar Y-chromosome background in cases and controls (see Supplementary Figure 3). This study was approved by the local ethics committees and all participants signed an informed consent.
Methods
Molecular analysis
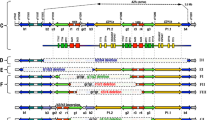
Genomic DNA was extracted from peripheral blood samples using a standard method.23 The screening for Yq microdeletions was performed according to the European Academy of Andrology (EAA)/European Molecular Genetics Quality Network (EMQN) guidelines24 with the addition of the STS sY1201 (Figure 1b). Detection and molecular characterization of partial AZFc rearrangements was performed according to a previously reported method.20 Briefly, we analyzed STSs sY1291, sY1191, sY1189, sY1197 and sY1192 (see GeneBank accessions G72340, G73809, GF102061, G67168 and G67166 for PCR primers and conditions) and identified the gr/gr deletion by the absence of sY1291 and sY1189 and b2/b3 deletion by the absence of sY1192 and sY1191.
Representation of the 21 ‘genuine’ Yq microdeletions detected. (a) Schematic representation of the Y chromosome showing the three AZF regions. (b) STS markers used for the diagnosis of Y microdeletions : in bold are the STSs used for the first step screening, and in italic are the ones used for the determination of the breakpoints of deletions (c) Type and number (n) of AZF deletions reported in the current study.
Y hgr definition
Y hgr was defined in all individuals with partial AZFc rearrangements and in 21 AZF classical deletion carriers. In addition, ∼60% of subjects recruited for the case/control study were analyzed in order to exclude population stratification bias. All individuals were genotyped for six binary markers (M145, M96, M9, M45, M168 and LLy22g) using a multiplexed primer set previously described25 and adapted for SNaPshot single base extension (Applied Biosystems, Foster City, CA, USA). This allowed the definition of eight hgr branches: A,B; DE,D; E; C,F,G,H,I; K,L,M,NO,O,S,T; J; N; P,Q,R.26 Marker 12f12 was tested only to discriminate between hrgs C,F,G,H,I and hgr J.
Statistical analysis
SPSS (version 17.0, Chicago, IL, USA) was used. We tested the significance of the observed difference in the incidence of partial AZFc deletions between patients and controls using Fisher’s exact test. As SC and TMC were not normally distributed, we used the nonparametric median test to compare median values of SC and TMC between individuals grouped according to DAZ copy number (CN; P<0.05 as statistically significant). Potential confounding factor, for partial AZFc deletions, were avoided by screening only individuals with no partial duplications and vice versa.
Results
Routine diagnostic screening: AZF deletions and karyotype anomalies
Among the 806 patients, 27 were found with a complete AZF deletion (3.3%). Karyotype anomalies were reported in a total of 58 patients with the most frequent anomaly represented by Klinefelter syndrome (for details see Supplementary Table 2). Out of 27 deletion carriers, 6 showed abnormal karyotype: (1) 4 cases with the entire Yq missing (three 46,XX male and one with 46,X,i(Yp)) and (2) 2 terminal AZFbc deletions (chrY.hg19:g.(19357589_22570359)_(58912042_?)del) (LOVD3 data base Variant ID: 0000021249)27 with breakpoint at P5 palindrome described at the karyotype analysis as idic(Yp). Among the 689 infertile patients with normal karyotype, 21 ‘genuine’ Y-chromosomal microdeletions were identified (3.0%): (1) 1 complete AZFa (chrY.hg19:g.(14328345_14607475)_(15132293_15603923)del) (Variant ID: 0000021250); (2) 20 AZFc: two terminal (chrY.hg19:g.(24524070_24872541)_(58912042_?)del) (Variant ID: 0000021251) and 18 b2/b4 (chrY.hg19:g.(24524070_24872541)_(25316578_28457316)del) (LOVD3 ID: chrY_000070) AZFc deletions (Figure 1c).
AZF deletion frequency: Estimating deletion frequencies according to the etiology showed a relatively higher frequency of deletions in the ‘idiopathic’ (4.2%, 19/454) compared with the ‘nonidiopathic’ group (2.4%, 8/325) (Table 1). In order to evaluate the impact of semen phenotype and etiology on the deletion frequency, we calculated the frequency for distinct semen categories belonging to different etiologic/sperm concentration groups (Table 2). The large majority of AZF deletion carriers (21/27) were AZ men, most of whom were ‘idiopathic’ (13/152; 8.5%). Also in this case, the etiology seems to play an important role, as the deletion frequency in the ‘nonidiopathic’ group was significantly lower (2.2%; P=0.037). The deletion frequency in Spanish ‘idiopathic’ infertile men was significantly lower compared with the non-Spanish men (2.7% versus 7.9%; P=0.018).
Genotype/phenotype correlation: Only 6 subjects (all AZFc deletion carriers) presented spermatozoa in their ejaculate, 5 with <1 × 106/ml and 1 with 1.2 × 106/ml. At least 3 semen analyses were performed for each individual over 1–2 years. Among patients with <1 × 106/ml, two (07-026, 06-192) displayed a nearly stable SC over time (both ∼0.01 × 106/ml), whereas more evident, although not significant, oscillations in the range of cryptozoospermia (CR) were observed in 06-012 (SC=0.095–0.044 × 106/ml), 08-039 (SC=0.04–0.250 × 106/ml) and 07-313 (SC=0.15–0.250 × 106/ml). A temporal trend for sperm number reduction was observed in the b2/b4 AZFc deletion carrier with >1 × 106/ml (09-067), who displayed a SC decrease from SOZ (1.6 × 106/ml) to CR (0.260 × 106/ml), respectively. In two SOZ patients, ICSI was performed with success using ejaculated spermatozoa and resulted in the birth of a healthy girl in both cases. No pregnancy was achieved in the other four cases.
AZ men with AZFc deletion had variable testicular phenotypes ranging from ‘pure’ or ‘mixed’ Sertoli cell-only syndrome (SCOS) to ‘pure’ bilateral hypospermatogenesis. TESE was performed in 11 patients and only 1 had spermatozoa (9.1% sperm recovery rate upon TESE).
The patient with complete AZFa deletion (sample 10-452) had pure bilateral SCO histology and no spermatozoa could be recovered upon TESE. Detailed genotype/phenotype description and ART results for AZF deletion carriers are reported in Table 3.
Partial AZFc rearrangements: case/control association study
gr/gr deletion: A conventional gr/gr deletion (chrY.hg19:g.(24876071_25505070)_(25505734_25316178)del) (LOVD3 DB-ID: chrY_000067) was found in 17 subjects (12 infertile patients and 5 normozoospermic controls). The deletion frequency between the two groups was statistically significant (12/302, 3.9% vs 5/359, 1.4%; OR=2.853; 95% CI=1.017–8.007; P=0.032; Table 4a).
b2/b3 deletion: This type of deletion (chrY.hg19:g.(24524070_24872541)_(24876071_25505070)del) (LOVD3 DB-ID: chrY_000068) was found only in the patient group (4/296) with a significantly different frequency compared with controls (0/354) (1.3% vs 0.0%, P=0.043).
A detailed description about gr/gr and b2/b3 deletion carriers is given in Supplementary Table 3.
Atypical deletions: Two patients (11-513 and 05-236) presented an atypical deletion pattern (both sY1291 and sY1191 positive) associated with the removal of DAZ3/4 and CDY1B (g.[26909216_27053187del;26191377_26194161del]) (variant ID: 0000021252) in the first subject and of DAZ3/4 and both CDY1 copies (g.[26909216_27053187del; 26191377_26194161del; 27768264_27771049del]) (variant ID: 0000021253) in the second subject.
All partial deletions: We calculated the combined frequency of all the deletions that decrease the AZFc gene content of at least 50%, observing a significantly higher frequency in patients compared with controls (5.8% vs 1.8%, respectively; OR=4.196; 95% CI=1.576–11.170; P=0.001; Table 4a).
AZFc duplications: an increased DAZ gene dosage (6 or 8 DAZ copies) associated with a simultaneously increased CDY1 gene dosage (3 or 4 copies) was observed in 28 Y chromosomes. These CNVs likely derive either from gr/gr (chrY.hg19:g.(24876071_25505070)_(25505734_25316178)dup) (Variant ID: 0000021256) or b2/b4 duplication (chrY.hg19:g.(24524070_24872541)_(25316578_28457316)dup) (Variant ID: 0000021258) events and did not show significant differences between patients (4.9%) and controls (3.5%) (Table 4a). We also found one control displaying a gr/gr deletion–duplication (chrY.hg19:g.[(24876071_25505070)_(25505734_25316178)del;(24524070_24872541)_(25316578_28457316)dup] (Variant ID: 0000021254) characterized by 4 CDY1B and 8 DAZ3/4 gene copies. Moreover, another patient carried a b2/b3 deletion followed by a b2/b4 duplication (chrY.hg19:g.[(24524070_24872541)_(24876071_25505070)del;(24524070_24872541)_ (25316578_28457316)dup] (Variant ID: 0000021255) that restored the reference gene dosage, and thus presented 4 DAZ copies (DAZ1/2 and DAZ3/4) and 2 copies of CDY1B.
Isolated CDY1 and DAZ CNVs: Two controls showed an isolated increase of CDY1 CN with 3 and 4 CDY1, respectively. Finally, isolated amplification of DAZ was found in 13 subjects: 10 (4 patients and 6 controls) with 6 DAZ copies and 3 (all controls) with 8 DAZ copies (Table 4b).
Impact of the DAZ CNVs on semen quality
In order to evaluate the effect of DAZ gene CNVs on semen quality, we grouped all subjects into five different categories: 0, 2, 4, 6 and 8 DAZ gene copies (for details see Table 4b). Men with 0 and 2 DAZ gene copies showed a significantly lower SC (median with 25th/75th percentiles: 0.0 × 106; 0.0–0.07 and 3.0 × 106/ml; 0.16–15.0, respectively) and TMC (0.0 × 106; 0.0–0.0 and 2.4 × 106; 0.13–30.0, respectively) compared with those bearing 4 DAZ gene copies (median SC 35.0 × 106/ml; 0.16–15.0 and median TMC of 31.3 × 106; 0.13–125.9). Increased DAZ gene CN (both 6 and 8 copies), although showing lower sperm count, was not significantly associated with reduced sperm quality (Figure 2).
DAZ gene copy number effect on sperm concentration (a) and total motile sperm count (TMC) (b) in the case/control study population. Data are presented as median (25th–75th percentiles). (a) Comparison between subjects with different DAZ gene copy numbers (DAZ CNs), showing significantly reduced sperm concentration in men with 0 and 2 DAZ copies versus 4 DAZ copies (4 DAZ copies are considered as a ‘normal’ gene dosage). (b) Men with 0 and 2 DAZ CNs have significantly lower TMC as compared with men with the ‘normal’ gene CNs (4 DAZ).
Effect of Y-chromosome background
Y hgr analysis was performed in order to: (1) search for a putative association between Y background and formation of complete AZF deletions and partial AZFc rearrangements; and (2) further evaluate the contribution of Y hgr to the phenotypic expression of the gr/gr deletion. Branches P,Q,R were the most represented in the whole study population (63.1% of all subjects analyzed; Supplementary Figure 1). All Spanish AZF deletion carriers belonged to these hgrs (Supplementary Figure 2A), which conversely made up only 15.4% of non-Spanish carriers. In fact, the Y hgr mostly observed in this cohort was hgr E (23.1%), found in 3 African patients (2 from Morocco and 1 from Cameroon), followed by the branches C,F,G,H,I; J (15.4% each) and K,L,M,NO,S,T/N (7.7%; Supplementary Figure 2B). Concerning the gr/gr deletion study (exclusively based on the Spanish population), we observed a similar Y hgr distribution in patients and controls: the majority belonged to the hgrs P,Q,R (63.5% and 62.7%, respectively), whereas the rest showed matching frequencies between the two groups (Supplementary Figure 3). Similarly, branches P,Q,R were the most represented among gr/gr deletion carriers in both patients (9/12; 75%) and controls (3/5; 60%) and none of the other Y hgrs observed (E; J; K,L,M,NO,S,T/N; C,F,G,H,I) showed a significant enrichment in the two phenotypic groups.
Discussion
Many aspects of Y-chromosome microdeletions have been clarified (mechanism of formation, identification of the genes involved, semen phenotype-dependent variation of the deletion frequency), but a few debated issues merit further discussion. First of all, it is unclear whether Y background might predispose to the formation of deletions, thus contributing to the observed ‘interpopulation’ variation in the deletion frequency. In our study, the significantly higher deletion frequency observed in non-Spanish compared with Spanish idiopathic infertile men is plausibly because of the different proportion of AZ men (44.4% in the non-Spanish versus 29.3% in the Spanish group); however, the Y background might also represent a contributory factor influencing deletion frequencies. Y hgr analysis showed, as expected, that Spanish AZF deletion carriers all belonged to the P,Q,R lineages. In the non-Spanish cohort, consistent with the high proportion of North African patients (53.2%) included, AZF deletions were mostly found on hgr E, which is seemingly more prone to Y microdeletions.28, 29 Moreover, the deletion frequency reported in idiopathic AZ and SOZ men (9.09 and 5.5%) in Moroccan population28 is consistent with our findings in non-Spanish idiopathic AZ and SOZ men (10.7% and 6.9%, respectively). The prevalence of Y microdeletions in our study population (3.3%) is in line with the overall data presented in the literature (3.5%, according to a recent meta-analysis30). When comparing our results with the German population (the lowest ever frequency in the literature1) for similar semen categories, we found slightly higher frequencies in our Spanish population. On the other hand, the deletion frequency in AZ Spanish and Italian population31 (displaying a more similar Y background) was almost identical (7.3% versus 7.2%), further supporting a possible Y-background effect on deletion frequencies.
Another debated issue concerns the sperm concentration cutoff value for routine diagnostic testing and more precisely whether Y-microdeletion screening should be indicated for all oligozoospermic men with <5 × 106/ml. By an in-depth literature review, we could observe that only 2.0% of all AZF deletion carriers with an explicitly indicated SC presented >2 × 106/ml (Supplementary Table 4). Accordingly, in our study only one carrier had >1 × 106/ml but the SC did not exceed 2 × 106/ml. These findings suggest that Y-microdeletion screening could be eventually restricted to infertile men with SC ≤2 × 106/ml. We found an AZFc deletion in two apparently non-idiopathic AZ patients: one (13-124) presenting with unilateral absence of vas deference and the other (07-339) with bilateral cryptorchidism. In both cases, the presence of the microdeletion, rather than the mere clinical condition, explains the AZ phenotype. This implies that the Yq screening in azoospermic men should be performed independently of the presence or absence of other abnormal andrological findings.
The predictive value of AZF deletions for sperm retrieval at TESE is also still debated. The majority of complete AZFa deletion carriers show SCOS in their testes; however, data are extremely scarce and the largest published review reported the presence of spermatids in the testes in 2/26 men.32 Our patient with the complete AZFa deletion showed a complete bilateral SCOS, further supporting that TESE should not be attempted in AZFa carriers. Our survey reports a sperm retrieval of 9.1% (1/11) in AZ men with AZFc deletion. This value lies below the lower limit of the range of sperm recovery rates reported in AZFc-deleted patients (14.3–80.0%).1, 31, 33, 34, 35, 36, 37 This is most likely related to technical issues such as low amount of testicular sample retrieved (single biopsy from each testis) and the procedure used (classical TESE); indeed, laboratories that performed microTESE reported higher sperm recovery (Supplementary Table 5). The high proportion of pure SCOS cases among our AZFc carriers represents another possible explanation for such a low retrieval rate.
Concerning the OZ subjects, there are studies both in favor and against the need to cryopreserve spermatozoa to counteract the progressive deterioration of sperm quality (from SOZ to AZ). We observed a single AZFc deletion carrier with SOZ that developed into CR, indicating that a progressive decline in spermatogenic activity in patients bearing AZFc deletions may occur. However, further longitudinal studies are needed to distinguish between physiological oscillations and real impairment of sperm parameters over time. Several authors proposed a higher risk for Turner syndrome and ambiguous genitalia in ICSI babies born from AZFc deletion carriers (for review see Simoni et al24). Our survey reveals two successful pregnancies with healthy female offspring, providing additional data to the presently scarce literature about this issue (44 babies described so far). As for partial AZFc rearrangements, one of the strengths of this study lies in the careful selection of patients and controls considering both phenotype (only strictly idiopathic infertile and normozoospermic controls were included) and geographic origin (all individuals were rigorously of Spanish ancestry). The Y-chromosome hgr analysis in patients and controls further demonstrated the lack of population stratification bias in the study. The selective recruitment strategy, together with the detailed molecular characterization of the AZFc region in the whole study population, allowed us to provide highly reliable data on both partial AZFc deletions and duplications.
Concerning the gr/gr deletion, we found that Spanish gr/gr deletion carriers have an increased probability (OR=2.8) of impaired spermatogenesis compared with noncarriers. Overall, these data together with a previous pilot study19 further confirm the gr/gr deletion as a significant risk factor in the Spanish population (OR=4.8; 95% CI=1.863–12.623; P<0.001; Supplementary Table 6), providing additional support of its clinical relevance in Caucasians, consistent with the meta-analyses published so far.14, 15, 16, 17 The clinical implication of this finding in the Spanish population reinforces the idea that the gr/gr deletion screening should gain more consideration when dealing with infertile couples. This issue is of particular importance considering that, in some populations, partial deletions were shown to favor the occurrence of complete deletions.38, 39
The majority of gr/gr deletion carriers belonged, as expected, to the P,Q,R branches in both patients and controls, supporting that the phenotypic variability of gr/gr deletion is independent of Y-chromosomal background in Europeans.40 Interestingly, we found b2/b3 deletion only in the patient cohort (4/296; 1.3%) and only one carrier belonged to hgr N and thus had the constitutive b2/b3 deletion. The remaining three b2/b3 deletion carriers belonged to hrgs C,F,G,H,I, which is a frequent Y hgr in Moroccan population for which b2/b3 deletion has been recently reported in association with male infertility.28, 29 The significant association observed indicates a Y background-dependent deleterious effect of b2/b3 on spermatogenesis.
Here, we present the first screening for partial AZFc duplications in a large series of patients and controls of Spanish origin. Our data, consistently with those regarding the Italian population,20 confirmed that both partial and complete AZFc duplications do not represent any risk for spermatogenic failure in the Caucasian population.
When considering the DAZ CN independently from the type of partial rearrangements (gr/gr, b2/b3 or ‘atypical’ deletion patterns, partial or complete AZFc duplications), we found that the reduced DAZ CN (0 and 2 copies) is associated with a significant reduction in semen quality in terms of SC and TMC as previously reported by Noordam et al.21 On the other hand, for the first time we showed that an increase (6 or 8 copies) with respect to the most common DAZ gene dosage (4 copies) does not significantly affect semen quality.
In conclusion, our 8-year experience together with the literature review allowed us to further clarify a number of debated issues concerning the routine Y-chromosome microdeletion screening: (1) the indication for routine Y-chromosome microdeletion screening may be eventually limited to subjects with ≤2 × 106/ml; (2) in azoospermic AZFc deletion carriers, classical TESE is associated with a low sperm recovery rate (9.1%), and therefore microTESE, which allows better outcomes, should be regarded as the best option for sperm retrieval in these patients; and (3) Y background could partially account for the differences in deletion frequency among populations. Finally, in our view, gr/gr deletion screening can be considered as part of the diagnostic work-up of idiopathic oligozoospermic men as it is a confirmed cofactor that contributes to impaired sperm production.41 On the contrary, in line with the Italian data, partial AZFc duplication is unlikely to be involved in the etiopathogenesis of spermatogenic impairment in Caucasian populations.
References
Simoni M, Tüttelmann F, Gromoll J, Nieschlag E : Clinical consequences of microdeletions of the Y chromosome: the extended Münster experience. Reprod Biomed Online 2008; 16: 289–303.
Vogt PH, Edelmann A, Kirsch S et al: Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol genet 1996; 5: 933–943.
Repping S, Skaletsky H, Lange J et al: Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am J Hum Genet 2002; 71: 906–922.
Cruger DG, Agerholm I, Byriel L, Fedder J, Bruun-Petersen G : Genetic analysis of males from intracytoplasmic sperm injection couples. Clin Genet 2003; 64: 198–203.
Krausz C, Quintana-Murci L, Barbaux S et al: A high frequency of Y chromosome deletions in males with nonidiopathic infertility. J Clin Endocrinol Metab 1999; 84: 3606–3612.
Raicu F, Popa L, Apostol P et al: Screening for microdeletions in human Y chromosome—AZF candidate genes and male infertility. J Cell Mol Med 2003; 7: 43–48.
Oliva R, Margarit E, Ballescá JL et al: Prevalence of Y chromosome microdeletions in oligospermic and azoospermic candidates for intracytoplasmic sperm injection. Fertil Steril 1998; 70: 506–510.
Martínez MC, Bernabé MJ, Gómez E et al: Screening for AZF deletion in a large series of severely impaired spermatogenesis patients. J Androl 2000; 21: 651–655.
Skaletsky H, Kuroda-Kawaguchi T, Minx PJ et al: The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 2003; 423: 825–837.
Jobling MA : Copy number variation on the human Y chromosome. Cytogenet Genome Res 2008; 123: 253–262.
Repping S, Skaletsky H, Brown L et al: Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat Genet 2003; 35: 247–251.
Lin YW, Hsu LC, Kuo PL et al: Partial duplication at AZFc on the Y chromosome is a risk factor for impaired spermatogenesis in Han Chinese in Taiwan. Hum Mutat 2007; 28: 486–494.
Krausz C, Chianese C, Giachini C, Guarducci E, Laface I, Forti G : The Y chromosome-linked copy number variations and male fertility. J Endocrinol Invest 2011; 34: 376–382.
Navarro-Costa P, Gonçalves J, Plancha CE : The AZFc region of the Y chromosome: at the crossroads between genetic diversity and male infertility. Hum Reprod Update 2010; 16: 525–542.
Visser L, Westerveld GH, Korver CM et al: Y chromosome gr/gr deletions are a risk factor for low semen quality. Hum Reprod 2009; 24: 2667–2673.
Tüttelmann F, Rajpert-De Meyts E, Nieschlag E, Simoni M : Gene polymorphisms and male infertility—a meta-analysis and literature review. Reprod Biomed Online 2007; 15: 643–658.
Stouffs K, Lissens W, Tournaye H, Haentjens P : What about gr/gr deletions and male infertility? Systematic review and meta-analysis. Hum Reprod Update 2011; 17: 197–209.
Rozen SG, Marszalek JD, Irenze K et al: AZFc deletions and spermatogenic failure: a population-based survey of 20,000 Y chromosomes. Am J Hum Genet 2012; 91: 890–896.
De Llanos M, Ballescà JL, Gázquez C, Margarit E, Oliva R : High frequency of gr/gr chromosome Y deletions in consecutive oligospermic ICSI candidates. Hum Reprod 2005; 20: 216–220.
Giachini C, Laface I, Guarducci E, Balercia G, Forti G, Krausz C : Partial AZFc deletions and duplications: clinical correlates in the Italian population. Hum Genet 2008; 124: 399–410.
Noordam MJ, Westerveld GH, Hovingh SE et al: Gene copy number reduction in the azoospermia factor c (AZFc) region and its effect on total motile sperm count. Hum Mol Genet 2011; 20: 2457–2463.
World Health Organization: WHO Laboratory Manual for the Examination of Human Semen and Semen-Cervical Mucus Interaction 4th edn Cambridge (UK): University Press, 1999.
Miller SA, Dykes DD, Polesky HF : A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16: 1215.
Simoni M, Bakker E, Krausz C : EAA/EMQN best practice guidelines for molecular diagnosis of y-chromosomal microdeletions. State of the art 2004. Int J Androl 2004; 27: 240–249.
Noordam MJ, Repping S : The human Y chromosome: a masculine chromosome. Curr Opin Genet Dev 2006; 16: 225–232.
Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF : New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res 2008; 18: 830–838.
Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT : LOVD v.2.0: the next generation in gene variant databases. Hum Mutat 2011; 32: 557–563.
Eloualid A, Rhaissi H, Reguig A et al: Association of spermatogenic failure with the b2/b3 partial AZFc deletion. PLoS One 2012; 7: e34902.
Imken L, El Houate B, Chafik A et al: AZF microdeletions and partial deletions of AZFc region on the Y chromosome in Moroccan men. Asian J Androl 2007; 9: 674–678.
Hofherr SE, Wiktor AE, Kipp BR, Dawson DB, Van Dyke DL : Clinical diagnostic testing for the cytogenetic and molecular causes of male infertility: the Mayo Clinic experience. J Assist Reprod Genet 2011; 28: 1091–1098.
Ferlin A, Arredi B, Speltra E et al: Molecular and clinical characterization of Y chromosome microdeletions in infertile men: a 10-year experience in Italy. J Clin Endocrinol Metab 2007; 92: 762–770.
Kleiman SE, Almog R, Yogev L et al: Screening for partial AZFa microdeletions in the Y chromosome of infertile men: is it of clinical relevance? Fertil Steril 2012; 98: 43–47.
Maurer B, Gromoll J, Simoni M, Nieschlag E : Prevalence of Y chromosome microdeletions in infertile men who consulted a tertiary care medical centre: the Münster experience. Andrologia 2001; 33: 27–33.
Peterlin B, Kunej T, Sinkovec J, Gligorievska N, Zorn B : Screening for Y chromosome microdeletions in 226 Slovenian subfertile men. Hum Reprod 2002; 17: 17–24.
Hopps CV, Mielnik A, Goldstein M, Palermo GD, Rosenwaks Z, Schlegel PN : Detection of sperm in men with Y chromosome microdeletions of the AZFa, AZFb and AZFc regions. Hum Reprod 2003; 18: 1660–1665.
Stahl PJ, Masson P, Mielnik A, Marean MB, Schlegel PN, Paduch DA : A decade of experience emphasizes that testing for Y microdeletions is essential in American men with azoospermia and severe oligozoospermia. Fertil Steril 2010; 94: 1753–1756.
Choi JM, Chung P, Veeck L, Mielnik A, Palermo GD, Schlegel PN : AZF microdeletions of the Y chromosome and in vitro fertilization outcome. Fertil Steril 2004; 81: 337–341.
Zhang F, Lu C, Li Z et al: Partial deletions are associated with an increased risk of complete deletion in AZFc: a new insight into the role of partial AZFc deletions in male infertility. J Med Genet 2007; 44: 437–444.
Lu C, Zhang J, Li Y et al: The b2/b3 subdeletion shows higher risk of spermatogenic failure and higher frequency of complete AZFc deletion than the gr/gr subdeletion in a Chinese population. Hum Mol Genet 2009; 18: 1122–1130.
Krausz C, Giachini C, Xue Y et al: Phenotypic variation within European carriers of the Y-chromosomal gr/gr deletion is independent of Y-chromosomal background. J Med Genet 2009; 46: 21–31.
Krausz C : Male infertility: pathogenesis and clinical diagnosis. Best Pract Res Clin Endocrinol Metab 2011; 25: 271–285.
Acknowledgements
We thank the patients for participating in this study, the clinicians of the Andrology Service (Drs G Ortiz and S Camarena) for providing samples, the staff of Seminology and Embriology Laboratory (Drs A Mata, O Lopez, A Garcia, O Martinez and R Gusta) for providing seminal phenotypes and information about ART outcomes and M Vieri and A Morales for technical support. A special thanks to Esperança Martí i Salís, president of Fundació Puigvert, for her continuous support. We thank the IIB Sant Pau-Fundació Puigvert Biobank for kindly providing some of the DNA samples. This work was funded by grants from the Spanish Health Ministry (FIS-08/1138 and FIS 11/02254) to CK and from the Spanish Ministry of Economy and Competitiveness (BFU 2009-07118) to RO.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Lo Giacco, D., Chianese, C., Sánchez-Curbelo, J. et al. Clinical relevance of Y-linked CNV screening in male infertility: new insights based on the 8-year experience of a diagnostic genetic laboratory. Eur J Hum Genet 22, 754–761 (2014). https://doi.org/10.1038/ejhg.2013.253
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2013.253
Keywords
This article is cited by
-
Detection of AZFc gene deletion in a cohort of Egyptian patients with idiopathic male infertility
Journal of Genetic Engineering and Biotechnology (2023)
-
High incidence of AZF duplications in clan-structured Iranian populations detected through Y chromosome sequencing read depth analysis
Scientific Reports (2023)
-
Characterizing the evolution and phenotypic impact of ampliconic Y chromosome regions
Nature Communications (2023)
-
Y chromosome structural variation in infertile men detected by targeted next-generation sequencing
Journal of Assisted Reproduction and Genetics (2021)
-
Molecular delineation of small supernumerary marker chromosomes using a single nucleotide polymorphism array
Molecular Cytogenetics (2020)