Abstract
The phenotype of patients with a chromosome 1q43q44 microdeletion (OMIM; 612337) is characterized by intellectual disability with no or very limited speech, microcephaly, growth retardation, a recognizable facial phenotype, seizures, and agenesis of the corpus callosum. Comparison of patients with different microdeletions has previously identified ZBTB18 (ZNF238) as a candidate gene for the 1q43q44 microdeletion syndrome. Mutations in this gene have not yet been described. We performed exome sequencing in a patient with features of the 1q43q44 microdeletion syndrome that included short stature, microcephaly, global developmental delay, pronounced speech delay, and dysmorphic facial features. A single de novo non-sense mutation was detected, which was located in ZBTB18. This finding is consistent with an important role for haploinsufficiency of ZBTB18 in the phenotype of chromosome 1q43q44 microdeletions. The corpus callosum is abnormal in mice with a brain-specific knock-out of ZBTB18. Similarly, most (but not all) patients with the 1q43q44 microdeletion syndrome have agenesis or hypoplasia of the corpus callosum. In contrast, the patient with a ZBTB18 point mutation reported here had a structurally normal corpus callosum on brain MRI. Incomplete penetrance or haploinsufficiency of other genes from the critical region may explain the absence of corpus callosum agenesis in this patient with a ZBTB18 point mutation. The findings in this patient with a mutation in ZBTB18 will contribute to our understanding of the 1q43q44 microdeletion syndrome.
Similar content being viewed by others
INTRODUCTION
The phenotype of patients with a submicroscopic deletion of chromosome 1q43q44 (OMIM: 612337) is characterized by intellectual disability, microcephaly, growth restriction, dysmorphic features, agenesis of the corpus callosum (ACC) and cardiac, gastroesophageal and urogenital anomalies.1, 2, 3, 4, 5, 6, 7, 8 Even for patients with an overlapping deletion, the phenotype varies considerably. So far, no direct association has been identified between the phenotype and the deleted genes. On the basis of overlapping microdeletions, a number of candidate genes have previously been suggested for microcephaly and corpus callosum agenesis. These include AKT3, ADSS, CEP170, Clorf100, Clorf101, Clorf121, Clorf199, EFCAB2, FAM36A, HNRNPU, HNRNPU-AS1, PLD5, PNAS-4, and SDCCA8.1, 3, 8, 9, 10, 11, 12, 13, 14 Specifically, ZBTB18 has repeatedly been identified as a strong candidate gene for microcephaly and/or ACC.7, 9, 11, 14, 15 ZBTB18 is particularly compelling since a brain-specific knock-out of this gene in mice causes microcephaly and callosal anomalies.16 However, other studies suggest critical regions that do not include ZBTB18.8, 10, 12
To our knowledge, no point mutations in ZBTB18 have been described so far. Here, we describe a patient with a de novo non-sense mutation in ZBTB18 and phenotypic features of the 1q43q44 microdeletion syndrome.
MATERIALS AND METHODS
Clinical features
This female was the first child of healthy, non-consanguineous parents. The pregnancy was established through in vitro fertilization, because of subfertility due to oligospermia. Amniocentesis, performed because of raised maternal serum markers, revealed a normal female karyotype. On ultrasound at 20 weeks of gestation bilateral pyelectasia and choroid plexus cysts were noted. Labor was induced at 36 weeks of gestation because of intra-uterine growth restriction.
Birth length was 45.5 cm (−2.3 SD), with a weight of 2.15 kg (−2.3 SD), and a head circumference of 30 cm (−2 SD). Neonatal examination was unremarkable. Ultrasound investigations of the brain and the abdomen showed no abnormalities.
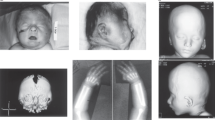
The girl developed feeding problems after the introduction of formula milk and weight gain was poor. After elimination of cow’s milk from the diet, her weight gain remained delayed. Gastroenterological examinations (including biochemistry, endoscopy, small bowel biopsy, and abdominal ultrasound and upper GI tract X-rays) revealed gastroesophageal reflux, but no other explanation for her growth restriction. Because of developmental delay, and suspected absence seizures associated with vomiting episodes, neurological assessment was performed which was unremarkable. Additional investigations included a cerebral MRI, EEG, metabolic screen, and hearing tests. All results were normal (Figure 1c).
Female with a de novo non-sense mutation in ZBTB18: c.397G>T (p.(Glu133*)). (a) Patient at age 16 months. Dysmorphic features include arched eyebrows with telecanthus, short palpebral fissures, a long nose with a prominent tip, short philtrum, and micrognathia; (b) Patient at age 34 months. Dysmorphic features are similar to those at age 16 months; however, the nasal tip is less prominent, the philtrum is longer, and the upper lip is thinner. (c) Lateral view of the brain at age 15 months. Note the presence of a normal corpus callosum (white arrow).
At the age of 16 months, she was referred to the genetics clinic for further assessment. At this time, all growth parameters were below −2.5 SD. Her height was 70.5 cm, weight 7 kg, and head circumference 41.5 cm. On clinical examination, she had proportionate short stature, mild facial dysmorphic features (Figure 1a and b), and slightly broad first fingers and toes. At later reviews, her delay became more evident and more marked in the speech domain. She walked unsupported at 26 months. At 34 months, her cognitive development was at the level of 2 years old with prominent speech delay. Her speech was hyper nasal. Her height was 79 cm (−4.5 SD), with a weight of 7.9 kg (−4.6 SD), and a head circumference of 43 cm (−4.1 SD).
At this time, a diagnosis of possible Floating-Harbor syndrome was considered.
Subsequent mutation analysis of the SRCAP gene revealed no abnormalities. MLPA assays for telomeric and recurrent genomic rearrangements (MRC-Holland kits) were also normal, as was a genomic microarray.
Molecular analysis
Exome sequencing was performed in a female patient from a cohort of patients with short stature, developmental delay, predominantly speech delay, with or without facial dysmorphic features. Exome sequencing was done on DNA from the female and both parents as described previously.17, 18 In brief, genomic DNA was isolated from blood with the use of a QIAgen DNA mini kit (QIAgen, Venlo, The Netherlands). Exomes were enriched using a SOLiD-optimized SureSelect human exome kit (Agilent v2, 50 Mb, Agilent Technologies Inc., Santa Clara, CA, USA), followed by sequencing performed using the 5500xl platform (Life Technologies, Carlsbad, CA, USA). After sequencing the trio, candidate de novo events were selected by excluding variants seen in either parent, whereas recessive models were evaluated using bi-allelic variants inherited from both parents. Candidate mutations were validated by conventional Sanger sequencing methods in DNAs of the patient and her parents (Supplementary Tables 1–3) and annotated according to HGVS recommendations. After publication, the variant will be submitted to the Human Gene Mutation Database (HGMD; URL: http://www.hgmd.cf.ac.uk/ac/index.php).
RESULTS
Exome sequencing detected three de novo variants. Two de novo missense variants were not confirmed with Sanger sequencing (Supplementary Table 3). Sanger sequencing confirmed the presence of a single de novo non-sense mutation located in ZBTB18: c.397 G>T (p.(Glu133*)) (NM_205768; UCSC, hg19). This mutation is predicted to cause a premature stop codon and may lead to non-sense mediated decay. No additional mutations in ZBTB18 were detected through Sanger sequencing in nine patients with an overlapping phenotype. Furthermore, neither stop mutation nor any other truncating event (stop-loss, frameshift mutation) in ZBTB18 was identified in 6500 individuals from the exome variant server (Exome Variant Server, NLHBI GO Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/) (accessed July 2013)).
DISCUSSION
We report a de novo non-sense mutation in ZBTB18 in a patient with global developmental delay, prominent speech delay, microcephaly, short stature, and discrete facial dysmorphisms. These clinical features are consistent with the phenotype of patients with microdeletions of chromosome 1q43q44 (Table 1). Overlapping clinical features include microcephaly (97%), developmental delay (100%), intra-uterine growth retardation (62%), and postnatal growth retardation (45%). We suggest that haploinsufficiency of ZBTB18 contributes to these features of the chromosome 1q43q44 microdeletion syndrome.
There are a number of reports on patients with deletions of 1q43q44 not including ZBTB18.1, 8, 9, 10, 11, 12 Out of 27 patients with deletions distal or proximal (including the AKT3 gene) to ZBTB18, only 5 (19%) showed corpus callosum abnormalities and 10 (37%) had microcephaly, compared with 82 and 97%, respectively, of patients with deletions including ZBTB18.1, 8, 9, 10, 11, 12 Furthermore, IUGR was seen in 3 out of 14 of these patients (21%) and postnatal growth retardation was seen in 7 out of 23 patients (30%). Collectively, these results support a crucial role for ZBTB18, but with contributions to the phenotype of other genes in patients with larger deletions.
ZBTB18, also known as ZNF238 or RP58, acts as a transcriptional repressor of key proneurogenic genes such as Neurogenin2 and NeuroD1.16 ZBTB18 is activated during neuronal differentiation in pin-like cells of the ventricular zone, and in migrating multipolar cells.19 ZBTB18 participates in neuron and astrocyte differentiation by mediating cell-cycle control of neural stem cells.20 Mice with loss of ZBTB18/RP58 die at birth with neocortical defects. CNS-specific loss of ZBTB18 is associated with microcephaly, ACC, and cerebellar hypoplasia.16 ZBTB18-mutant brains maintain precursor pools, but have reduced neuronal and increased glial differentiation. This is consistent with a role for ZBTB18 in favoring neuronal differentiation and brain growth by repressing multiple proneurogenic genes in a timely manner. On the basis of the phenotypes observed in mutant mice, Xiang et al16 proposed ZBTB18 as a strong candidate gene for the 1qter deletion syndrome. Our finding of intellectual disability and microcephaly in a female with a de novo non-sense mutation in ZBTB18 strongly supports this hypothesis.
Dysmorphic features in our female were mild, whereas variable facial features have been previously reported with 1q43q44 microdeletions of different sizes and gene content. Clearly, other genes from the 1q43q44 region may still contribute to the phenotypic outcome in patient cases with the 1q43q44 microdeletion syndrome. Most (24 out of 29) previously reported patients with a 1q43q44 microdeletion including ZBTB18 had corpus callosum abnormalities on brain imaging. The majority (66%) developed seizures.1, 8, 9, 10, 11, 12, 15 Incomplete penetrance is a possible explanation for the absence of ACC in this girl, given that the corpus callosum was apparently normal in at least five patients with 1q43q44 microdeletions that included ZBTB18 as described previously.1, 9, 12 We note that brain-specific loss of ZBTB18 in mice leads to a small brain phenotype with ACC and cerebellar hypoplasia.16
In conclusion, we describe the first patient with a de novo non-sense mutation in ZBTB18. The phenotype of the patient reported here suggests that ZBTB18 haploinsufficiency contributes to the microcephaly and global developmental delay that are core features of the 1q43q44 microdeletion syndrome.
References
Boland E, Clayton-Smith J, Woo VG et al: Mapping of deletion and translocation breakpoints in 1q44 implicates the serine/threonine kinase AKT3 in postnatal microcephaly and agenesis of the corpus callosum. Am J Hum Genet 2007; 81: 292–303.
De Vries BB, Knight SJ, Homfray T, Smithson SF, Flint J, Winter RM : Submicroscopic subtelomeric 1qter deletions: a recognisable phenotype? J Med Genet 2001; 38: 175–178.
Hill AD, Chang BS, Hill RS et al: A 2-Mb critical region implicated in the microcephaly associated with terminal 1q deletion syndrome. Am J Med Genet A 2007; 143A: 1692–1698.
Merritt JL 2nd, Zou Y, Jalal SM, Michels VV : Delineation of the cryptic 1qter deletion phenotype. Am J Med Genet A 2007; 143: 599–603.
Roberts AE, Cox GF, Kimonis V, Lamb A, Irons M : Clinical presentation of 13 patients with subtelomeric rearrangements and a review of the literature. Am J Med Genet A 2004; 128A: 352–363.
Rossi E, Piccini F, Zollino M et al: Cryptic telomeric rearrangements in subjects with mental retardation associated with dysmorphism and congenital malformations. J Med Genet 2001; 38: 417–420.
van Bever Y, Rooms L, Laridon A et al: Clinical report of a pure subtelomeric 1qter deletion in a boy with mental retardation and multiple anomalies adds further evidence for a specific phenotype. Am J Med Genet A 2005; 135: 91–95.
van Bon BW, Koolen DA, Borgatti R et al: Clinical and molecular characteristics of 1qter microdeletion syndrome: delineating a critical region for corpus callosum agenesis/hypogenesis. J Med Genet 2008; 45: 346–354.
Ballif BC, Rosenfeld JA, Traylor R et al: High-resolution array CGH defines critical regions and candidate genes for microcephaly, abnormalities of the corpus callosum, and seizure phenotypes in patients with microdeletions of 1q43q44. Hum Genet 2012; 131: 145–156.
Caliebe A, Kroes HY, van der Smagt JJ et al: Four patients with speech delay, seizures and variable corpus callosum thickness sharing a 0.440 Mb deletion in region 1q44 containing the HNRPU gene. Eur J Med Genet 2010; 53: 179–185.
Nagamani SC, Erez A, Bay C et al: Delineation of a deletion region critical for corpus callosal abnormalities in chromosome 1q43-q44. Eur J Hum Genet 2012; 20: 176–179.
Thierry G, Beneteau C, Pichon O et al: Molecular characterization of 1q44 microdeletion in 11 patients reveals three candidate genes for intellectual disability and seizures. Am J Med Genet A 2012; 158A: 1633–1640.
Speevak MD, Zeesman S, Leonard N, Nowaczyk MJ : Further evidence that a 100 Kb critical region is responsible for developmental delay, seizures, and dysmorphic features in 1q43q44 deletion patients. Am J Med Genet A 2013; 161A: 913–915.
Orellana C, Rosello M, Monfort S et al: Corpus callosum abnormalities and the controversy about the candidate genes located in 1q44. Cytogenet Genome Res 2009; 127: 5–8.
Perlman SJ, Kulkarni S, Manwaring L, Shinawi M : Haploinsufficiency of ZNF238 is associated with corpus callosum abnormalities in 1q44 deletions. Am J Med Genet A 2013; 161A: 711–716.
Xiang C, Baubet V, Pal S et al: RP58/ZNF238 directly modulates proneurogenic gene levels and is required for neuronal differentiation and brain expansion. Cell Death Differ 2012; 19: 692–702.
de Ligt J, Willemsen MH, van Bon BW et al: Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 2012; 367: 1921–1929.
Vissers LE, de Ligt J, Gilissen C et al: A de novo paradigm for mental retardation. Nat Genet 2010; 42: 1109–1112.
Ohtaka-Maruyama C, Hirai S, Miwa A, Takahashi A, Okado H : The 5′-flanking region of the RP58 coding sequence shows prominent promoter activity in multipolar cells in the subventricular zone during corticogenesis. Neuroscience 2012; 201: 67–84.
Hirai S, Miwa A, Ohtaka-Maruyama C et al: RP58 controls neuron and astrocyte differentiation by downregulating the expression of Id1-4 genes in the developing cortex. EMBO J 2012; 31: 1190–1202.
Acknowledgements
We would like to thank the patients and their parents. We also thank Bert B de Vries and David A Koolen for their clinical support. We thank the Genomic Disorders Group Nijmegen for the technical support in performing the whole-exome sequencing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
de Munnik, S., García-Miñaúr, S., Hoischen, A. et al. A de novo non-sense mutation in ZBTB18 in a patient with features of the 1q43q44 microdeletion syndrome. Eur J Hum Genet 22, 844–846 (2014). https://doi.org/10.1038/ejhg.2013.249
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2013.249
Keywords
This article is cited by
-
The mouse model of intellectual disability by ZBTB18/RP58 haploinsufficiency shows cognitive dysfunction with synaptic impairment
Molecular Psychiatry (2023)
-
Clinical and molecular characterization of 1q43q44 deletion and corpus callosum malformations: 2 new cases and literature review
Molecular Cytogenetics (2022)
-
Translational profiling of mouse dopaminoceptive neurons reveals region-specific gene expression, exon usage, and striatal prostaglandin E2 modulatory effects
Molecular Psychiatry (2022)
-
Cytogenomic characterization of 1q43q44 deletion associated with 4q32.1q35.2 duplication and phenotype correlation
Molecular Cytogenetics (2018)
-
Genetic and phenotypic dissection of 1q43q44 microdeletion syndrome and neurodevelopmental phenotypes associated with mutations in ZBTB18 and HNRNPU
Human Genetics (2017)