Abstract
Hereditary hearing loss is genetically heterogeneous, with a large number of genes and mutations contributing to this sensory, often monogenic, disease. This number, as well as large size, precludes comprehensive genetic diagnosis of all known deafness genes. A combination of targeted genomic capture and massively parallel sequencing (MPS), also referred to as next-generation sequencing, was applied to determine the deafness-causing genes in hearing-impaired individuals from Israeli Jewish and Palestinian Arab families. Among the mutations detected, we identified nine novel mutations in the genes encoding myosin VI, myosin VIIA and myosin XVA, doubling the number of myosin mutations in the Middle East. Myosin VI mutations were identified in this population for the first time. Modeling of the mutations provided predicted mechanisms for the damage they inflict in the molecular motors, leading to impaired function and thus deafness. The myosin mutations span all regions of these molecular motors, leading to a wide range of hearing phenotypes, reinforcing the key role of this family of proteins in auditory function. This study demonstrates that multiple mutations responsible for hearing loss can be identified in a relatively straightforward manner by targeted-gene MPS technology and concludes that this is the optimal genetic diagnostic approach for identification of mutations responsible for hearing loss.
Similar content being viewed by others
Introduction
Hundreds of mutations in more than 60 genes are known to be involved in hearing loss,1 yet it is estimated that for a majority of individuals with suspected hereditary hearing loss, the etiology remains unknown. In many of these unsolved cases, the causative mutation might be a known or novel mutation in a known gene that is not screened routinely due to practical limitations. Until recently, the large reservoir of known deafness genes, as well as their large size, precluded comprehensive genetic diagnosis. The combination of targeted genomic capture and massive parallel sequencing (MPS) has become a promising tool for detecting novel and known mutations involved in hereditary hearing loss and for solving many deafness cases in a rapid and cost-effective manner.2, 3 In our study, we applied this genomic method to determine the causative genes and mutations in undiagnosed deaf individuals of 96 Israeli Jewish and Palestinian Arab families. Among the mutations detected, we identified nine novel mutations in the genes encoding myosin VI, myosin VIIA and myosin XVA.
Myosins, a superfamily consisting of over 35 classes,4 are actin-based molecular motors. The N-terminal region of their heavy chain contains the most conserved sequence with the characteristic head domain, followed by a more variable neck region. The N-terminal, most conserved globular motor or head domain, binds actin and ATP. The neck or regulatory domain contains a variable number of IQ motifs that bind calmodulin or calmodulin-like light chains and amplifies the conformational changes of the motor domain as a lever arm to produce force. The C-terminal region of their heavy chain corresponds to the tail domain and is the most variable region between myosin classes. This diversity is responsible for specific myosin function such as filament formation, cargo binding and targeting to specific membrane proteins.5 Myosins are classified into two-headed conventional myosins, also referred to as myosins II, which form filaments and are the motors that power muscle contraction as well as cytokinesis. The other classes correspond to unconventional myosins, which can either be one- or two-headed myosins. Of the latter, several myosins have proven to have a crucial role in the inner ear.6 The evidence is very strong that mutations in four unconventional myosins, myosin IIIA7 (gene symbol MYO3A; recessive hearing loss DFNB30), myosin VI8, 9 (MYO6; dominant and recessive hearing loss DFNA22 and DFNB37, respectively), myosin VIIA10, 11 (MYO7A; dominant and recessive hearing loss DFNA11 and DFNB2, respectively) and myosin XVA12 (MYO15A; recessive hearing loss DFNB3), and two non-muscle conventional myosins, myosin, heavy chain 9, non-muscle13 (MYH9; dominant hearing loss DFNA17) and myosin, heavy chain 14, non-muscle14 (MYH14; dominant hearing loss DFNA4) lead to non-syndromic hearing loss. Mutations in three additional unconventional myosins, myosin IA15 (MYO1A; dominant hearing loss DFNA48), myosin 1C (MYO1C) and myosin 1F (MYO1F) have been associated with dominant non-syndromic hearing loss.16 MYO7A mutations are also responsible for Usher syndrome type IB17 and MYO6 mutations may be linked to cardiomyopathy as well.18 The type of hearing loss is quite variable among the affected individuals, with no clear genotype–phenotype correlation.
The mouse has been an instrumental model in the identification of human loci, helping to define myosin function in the inner ear. The shaker 1 mouse had a key role in the discovery of the genetic basis of Usher syndrome type IB, a form of retinitis pigmentosa associated with deafness.19 The first Myo6 mutation was detected in the deaf Snell's waltzer mouse.20 MYO15A mutations were discovered concomitantly in human DFNB3 families and in the shaker 2 mouse.12, 21 In the Middle Eastern Jewish and Arab populations, to date, eight mutations have been reported in myosins IIIA,7 myosin VIIA22 and myosin XVA.3, 22 The present study, using targeted capture and MPS, defines nine novel mutations in MYO6, MYO7A and MYO15A, doubling the number of myosin mutations present in the Middle East responsible for non-syndromic hearing loss.
Materials and methods
Family ascertainment
The study was approved by the Helsinki Committees of Tel Aviv University, the Israel Ministry of Health and the Human Subjects Committee of Bethlehem University. A total of 48 Israeli Jewish and 48 Palestinian Arab probands and their affected and unaffected relatives were ascertained for the study, as well as 200 Israeli Jewish and 263 Palestinian Arab unrelated hearing-impaired individuals. In addition, 600 and 100 Israeli Jewish and Palestinian Arab hearing controls, respectively, were included in the study. These additional probands and controls were screened for each mutation identified by MPS for determining the prevalence among the deaf population and carrier rates among the hearing population. A medical history was collected from each proband, including type and degree of hearing loss, age at onset, evolution and symmetry of the hearing impairment, use of hearing aids, presence of tinnitus, medication, noise exposure, pathologic changes in the ear and other relevant clinical manifestations, including vision problems and motor development, family history and consanguinity. Vestibulo-ocular reflex and fundoscopy were not examined. The inclusion criterion was idiopathic hearing loss with an emphasis on those with family history or consanguinity, although some singleton cases were included. Blood was drawn and genomic DNA extracted, after all subjects signed committee-approved consent forms.
Gene exclusion
All subjects were tested for GJB223 by Sanger sequencing. The remaining 12 deafness genes found in Jewish populations have a low prevalence, and thus known mutations were screened only in cases where subjects manifested a relevant phenotype or belonged to a relevant ethnic background. These genes include GJB6, PCDH15, USH1C, MYO3A, SLC26A4, LOXHD1, CDH23, MYO15A, WFS1, TECTA, POU4F3 and the inverted duplication of TJP2.3, 23, 24, 25 All known deafness-causing mutations in the Palestinian population were excluded, including mutations in CDH23, MYO7A, MYO15A, OTOF, PJVK, SLC26A4, TECTA, TMHS, TMPRSS3, OTOA, PTPRQ and GPSM2.22, 26, 27
Massive parallel sequencing
Capture libraries were created and MPS was performed, followed by bioinformatics analysis, as previously described.28, 29 Exons and the flanking 40 bp into the introns of 284 human deafness-associated genes were captured using the SureSelect Target Enrichment Solution (Agilent, Santa Clara, CA, USA) and sequenced on an Illumina HiSeq 2000 Analyzer (HT-Seq Unit, Technion, Haifa, Israel). To identify variants, reads were aligned to the Genome Reference Consortium Human Build 37 (GRCh37/hg19) using Burrows–Wheeler Aligner.30 Single-nucleotide variants (SNV) and insertion–deletions (indels) were called from the aligned data using SAMtools (http://samtools.sourceforge.net) and Dindel,31 respectively. Copy number variants (CNV) analysis was performed using BreakDancer.32 Rare variants were identified by filtering using the data from dbSNP135, the 1000 Genomes Project and the Exome Variant Server (http://evs.gs.washington.edu/EVS/) and classified by predicted protein effects using Polyphen2,33 SIFT,34 MutationTaster35 and Consurf.36 The novel variants were checked using Mutalyzer (http://www.LOVD.nl/mutalyzer/) and variants were uploaded to the LOVD locus-specific database (www.lovd.nl/MYO6; www.lovd.nl/MYO7A; www.lovd.nl/MYO15A).
RNA isolation
RNA was isolated from 5 ml of peripheral blood using the RNeasy Mini Kit (Qiagen, Hilden, Germany). One microgram of the extracted RNA was used as a template for reverse transcription using the qScript cDNA Synthesis Kit (Quanta Biosciences, Gaithersburg, MD, USA). PCR was performed on cDNA for the MYO6 gene using primers Exon9-F GCTCAGTTGTTGGAGGATTTG and Exon11-R CTGCCAGCTTCCTCAAAATC for 35 cycles.
Modeling of the mutations
The consequences of the novel mutations in myosin VI were inferred from the model of the structure of the myosin VI motor domain (PDB 2BKH). For the myosin VIIa mutations found in the motor domain, the different structures known for myosin Va (PDB 1OE,37 1 W7j38) were used.
Results
A targeted capture pool, previously developed for the identification of mutations in 284 known deafness genes, was used.28, 29 This pool comprises 121 human and 163 orthologues of mouse genes, including three microRNAs.
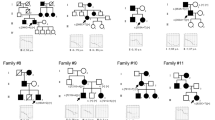
Two novel MYO6 mutations were detected in Palestinian Arab families (Table 1). Family AV presented with both moderate progressive late-onset and congenital profound bilateral hearing loss (Figure 1a). AV-III-1 is the daughter of a consanguineous marriage. Her auditory brainstem response results for alternative click stimuli showed no response at 90 dBnHL on both sides, at the age of 1 year, indicating profound deafness. The heterozygote paternal uncle of this child, AV-II-1, manifested moderate HL that was diagnosed in his mid thirties, and by the age of 53 his audiogram showed pure tone thresholds of 50–60 dB. The parents of AV-III-1 are heterozygote; the father did not exhibit a known hearing loss before his death at age 30 and the mother, at age 33, has a hearing impairment by family report. No candidate variants were detected in the indel and CNV analysis. The only homozygous variant candidate recovered in the SNV analysis was c.897G>T in the MYO6 gene (NM_004999.3), on chr6:76554694 in 161/165=98% reads. Sanger sequencing confirmed the presence of this variant. c.897G>T is a splice mutation, located in the last nucleotide of exon 10 of the MYO6 gene, leading to skipping of this exon (Figure 1c). This mutation was not detected among 263 deaf Palestinian probands or 100 hearing controls of Palestinian Arab origin.
Analysis of myosin VI mutations. (a) Pedigrees and segregation analysis of Family AV. (b) Partial sequences of MYO6 exon 10 from a normal hearing and affected proband AV-III-1 demonstrating the c.897G>T splicing mutation. (c) cDNA was amplified from blood from the patients of Family AV, using primers from exon 9–12 to generate a 545-bp fragment. Exon 10 was lost in the resultant cDNA of patient AV-III-1, confirmed by sequencing. (d) Pedigrees and segregation analysis of Family QS009. (e) Partial sequences of MYO6 from an affected proband demonstrating the c.2777T>A, p.Leu926Gln, as shown by a chromatogram. (f) Schematic representation of myosin VI shows the location of the potential splicing mutation c.897G>T in the ATPase activity domain in the motor and p.Leu926Gln, located in a potential proximal dimerization region in the tail of the myosin VI protein.
The second MYO6 mutation was detected in the Palestinian Arab family QS009, with a recessive mode of inheritance, who presented with congenital profound bilateral hearing loss (Figure 1d). The parents are consanguineous, with a total of six children, three of whom are profoundly deaf and three who have normal hearing. Indels and CNVs were excluded and the only homozygous variant candidate found in the SNV analysis was c.2777T>A in exon 26, at chromosomal position chr6:76599892 with 311/318=98% reads. Sanger sequencing revealed that both parents and one of the hearing brothers are heterozygous for the mutation, and the two deaf sisters are homozygous. No SNV was detected at this position in the Exome Variant Server. The c.2777T>A variant was confirmed in the proband by Sanger sequencing (Figure 1e). This mutation is predicted to create a missense mutation, p.(Leu926Gln) (NP_004990.3), located in a potential proximal dimerization region in the tail region of the myosin VI protein (Figure 1f). This variant is predicted to be damaging by Polyphen2 (0.79), SIFT (0) and MutationTaster (P=0.99). c.2777T>A was not detected among 263 deaf Palestinian probands or 100 hearing controls of Palestinian Arab origin.
Five novel mutations were identified in the MYO7A gene (NM_000260.3) in both Jewish and Arab populations (Table 1). Family D79 (Figure 2a) is an Israeli Jewish family with recessive inheritance originating from Turkey. The proband, with congenital profound hearing loss, is compound heterozygous for two variants in MYO7A. The first variant, corresponding to MYO7A c.29T>C at chromosomal position chr11:76853765, was supported by 9/26=37% of reads (Figure 2b). The predicted missense mutation, p.(Val10Ala) (NP_000251.3), is located at the beginning of the myosin head domain (Figure 2j) and is predicted to be damaging by PolyPhen2 (0.917) and MutationTaster (P=0.99) and supported by a ConSeq value of 8, indicating high conservation at this site. The second MYO7A variant, c.1969C>T in exon 17 at chr11:76885835 had 38/76=50% reads (Figure 2b). Similar to p.Val10Ala, the predicted p.(Arg657Trp) variant corresponds to a missense mutation in the myosin head of the myosin VIIA protein (Figure 2i and j). p.Arg657Trp is predicted to cause damage by Polyphen2 (0.999), SIFT (0) and MutationTaster (P=0.99) and supported by the highest ConSeq value of 9. p.Arg657Trp was superimposed on a structural model of the myosin VIIa motor domain (Figure 2i). Both variants were confirmed in proband D79-II-1 by sequencing the corresponding exons (Figure 2b). c.29T>C and c.1969C>T were present in a heterozygous state in the mother D79-I-2 and father D79-I-1, respectively. c.29T>C was not detected among 200 Jewish deaf probands or 221 normal hearing controls, including 112 of Turkish origin; c.1969C>T was identified in one Ashkenazi proband (T275), in a heterozygous state, out of 169 Jewish deaf probands tested. Among a Turkish control group, 2/133 (1.5%) were heterozygous carriers for the mutation, whereas 100 Ashkenazi hearing individuals and 77 controls of other ethnic groups were all wild type for the variant.
Analysis of myosin VIIA mutations. (a) Pedigrees and segregation analysis of Family D79. (b) Partial sequences of MYO7A from a normal hearing and affected proband D79-II-1 demonstrating the c.29T>C and c.1969C>T mutations. (c) Pedigrees and segregation analysis of Family D75. (d) Partial sequences of MYO7A from an affected proband demonstrating the c.620A>G mutation. (e) Pedigrees and segregation analysis of Family QS025. (f) Chromatogram of the c.6211C>T mutation. (g) Pedigrees and segregation analysis of families QS004 and QS013. (h) Chromatogram of the c.4153-2A>G mutation. (i) Myosin VIIA mutations p.Asn207Ser and p.Arg657Trp are superimposed on a structural model of the myosin VIIa motor domain, based on the structure of myosin V bound to ATP.38 Note in black the location of the p.Asn207Ser mutation that may result in poor nucleotide binding and hydrolysis. The p.Arg657Trp mutation is likely to impair the communication between the active site and the lever arm as it is located at the interface between two connectors of the motor relay (yellow) and the SH1 helix (red) that controls the rotation of the lever arm. (j) Schematic representation of the myosin VIIA protein shows the location of p.Val10Ala, p.Arg657Trp, p.Asn207Ser, p.Gln2071* and c.4153-2A>G.
A third MYO7A mutation was detected in Jewish family D75 originating from Uzbekistan (Figure 2c), with recessive congenital profound hearing loss. After excluding all indel and CNV variants, the only homozygous variant candidate revealed in the SNV analysis was c.620A>G in exon 7 at chr11:76867935, supported by 400/409=98% reads and confirmed in the proband by Sanger sequencing (Figure 2d). Both parents were found to be heterozygous for c.620A>G by Sanger sequencing. This predicted p.(Asn207Ser) variant also corresponds to a missense mutation located in the myosin head of the myosin VIIA protein (Figure 2i and j). p.Asn207Ser can easily be modeled in the structure of a myosin bound to MgATP (such as the post-rigor state of myosin Va: 1w7j) (Figure 2i). The model shows that the Asn residue is critical for the binding of the nucleotide and that the serine side chain cannot perform a similar function, predicting a much lower affinity of the motor for the nucleotide (Figure 2i). c.620A>G was detected among 3/171 Jewish deaf probands in a heterozygous state, all of Ashkenazi origin. One of these three heterozygous probands, T275, was also found to be heterozygous for the c.620A>G mutation, that is, T275 is compound heterozygous for the two novel myosin VIIA p.Asn207Ser/p.Arg657Trp mutations. As the parents’ DNA was unobtainable, we were unable to confirm that the mutations are in trans. Interestingly, another proband heterozygous for the myosin VIIA p.Asn207Ser mutation, D114, is also heterozygous for the PCDH15 p.Arg245* mutation. p.Asn207Ser was not detected among 266 normal hearing controls, including 78 of Kavkazian origin, 100 Ashkenazi and 88 other mixed ethnicities.
An additional MYO7A mutation was detected in Palestinian Arab family QS025 with no family history of deafness, with QS025-II-1 with congenital profound hearing loss (Figure 2e). All indels and CNVs were excluded and the only homozygous SNP candidate was the MYO7A c.6211C>T variant at chromosomal location chr11:76922356, with 37/37=100% reads, confirmed by Sanger sequencing in the proband (Figure 2f). The mother was found to be heterozygous for the mutation. The predicted p.(Gln2071*) mutation, located in the FERM 2 domain of the tail of the myosin VIIA protein (Figure 2j), creates a stop codon at exon 45 (out of 49), predicted to lead to a truncated protein. p.Gln2071* was not detected among 263 deaf Palestinian probands or 100 hearing controls of Palestinian Arab origin.
The fifth MYO7A mutation was identified in two Palestinian Arab families, QS004 and QS013, with congenital profound deafness inherited in a recessive mode. QS004-II-1, a member of a consanguineous family, has two affected and two unaffected siblings (Figure 2g). QS013-II-1 is a member of a consanguineous family, with two out of seven children affected. After excluding all indel and CNV variants, the only homozygous variant candidate revealed in the SNV analysis for both patients was c.4153-2A>G at chr11:76905397, with 97/98=99% (QS004-II-1) and 67/68=99% reads (QS013-II-1). In both cases, the mutation segregated with hearing impairment in the families (Figure 2h). c.4153-2A>G, located in the FERM 1 domain of the tail of myosin VIIA protein (Figure 2j), is positioned two nucleotides upstream of exon 32 and abolishes the consensus AG 3’ splice site. As the cDNA could not be amplified by PCR, the putative splice mutation was analyzed by the Alternative Splice Site Predictor (ASSP) tool (http://wangcomputing.com/assp/index.html) for splice-site damage, demonstrating that no splicing can occur at this position with the c.4153-2A>G mutation. The mutation was not detected among 263 deaf Palestinian probands or 100 hearing controls of Palestinian Arab origin.
A novel MYO15A mutation was detected in Palestinian Arab family BA, with congenital profound deafness affecting both siblings in a recessive mode, in a compound heterozygous state with one known MYO15A c.4240G>A mutation3 (Figure 3a) present with 99/210=47% reads. The second variant c.1223C>T was identified by MPS, after indels and CNVs were excluded, in exon 2 (NM_016239.3) at chromosomal location chr17:18023337 with 177/368=48% reads. Both mutations were confirmed in the proband by Sanger sequencing and segregated with hearing loss in the family (Figure 3a). The p.(Ala408Val) (NP_057323.3) variant, located in the N-terminal extension before the motor domain of the myosin XVA protein (Figure 3e), is predicted to be possibly damaging by Polyphen2 (0.445), although benign by SIFT (0.46). It was reported in 0.0018 of the 1000 Genome population and appeared in the SNP database (rs191710555), with an unknown validation status or clinical significance (http://www.ncbi.nlm.nih.gov/projects/SNP/). This SNP was not reported in the Exome Variant Server. Although MutationTaster suggests that this variant is a polymorphism (referring to the SNP database), it also states that the amino-acid change might affect protein features and may result in splice-site changes. The c.1223C>T variant was not detected among 263 deaf Palestinian probands or 100 hearing controls of Palestinian Arab origin.
Analysis of myosin XVA mutations. (a) Pedigree and segregation analysis of Family BA. (b) Partial sequence of MYO15A from an affected proband BA-II-1 demonstrating the compound heterozygote mutation c.1223C>T, p.Ala408Val, which appeared with the known p.Glu1414Lys mutation. (c) Pedigree and segregation analysis of Family 1234. (d) Partial sequences of MYO15A from an affected proband 1234-II-1 demonstrating the mutation c.8467G>A, p.Asp2823Asn. (e) Schematic representation of the myosin XVA protein shows the location of the p.Ala408Val, p.Glu1414Lys and p.Asp2823Asn mutations.
The second MYO15A mutation was identified in Israeli Jewish family 1234, originating from Iran (Figure 3c), with a recessive mode of inheritance and congenital profound hearing loss. c.8467G>A in exon 47 at chromosomal position chr17:18059516 was detected by MPS with 170/173=98% reads and was confirmed by Sanger sequencing (Figure 3d). The predicted variant, p.(Asp2823Asn), is located in the tail domain of the myosin XVA protein (Figure 3e) and is predicted to be damaging by Polyphen2 (0.977), SIFT (0) and MutationTaster (P=0.99). A ConSeq value of 8 suggests high conservation at this location. c.8467G>A was detected in one out of 216 Israeli Jewish deaf probands, D36C, the son of a consanguineous marriage, with both parents originating from Iran, in a homozygous state, bringing the prevalence of the mutation among the deaf Jewish population tested to 0.9% (4/432 alleles). Among 349 Jewish normal hearing controls, including 171 of Iranian origin, two carriers were detected, both of Iranian origin, resulting in a carrier rate of 1.2% (2/171) in the Jewish Iranian population tested.
Discussion
Targeted enrichment and MPS of human and mouse deafness genes has proven to be an efficient screening tool for deafness mutations.2, 3 Our goal is to utilize this method in order to help solve the molecular etiology of the deaf population in the Middle East. Routine diagnostic tests conducted in the clinic, most of which do not use MPS, have the disadvantage of overlooking known and novel mutations in known deafness genes, some of which are very common in other populations. As a considerable number of these genes are comprised of multiple exons, it is unrealistic, in terms of time and cost, to screen for them individually in the clinic. These large genes include the myosins, a family of proteins in which dozens of mutations have been reported worldwide (Deafness Variation Database, http://deafnessvariationdatabase.org), with only a few previously found in the Middle East. MYO6 (NM_004999.3), with 35 exons, encodes a 1285-amino-acid protein, myosin VI; MYO7A (NM_000260.3), containing 49 exons, encodes a protein of 2215 amino acids, myosin VIIA; and MYO15A (NM_016239.3), with 65 exons, encodes a protein of 3530 amino acids, myosin XVA. Together, they encompass 149 exons for only three genes, which are impractical to sequence by conventional Sanger sequencing. Using a combined approach of targeted gene capture and deep sequencing, we have identified nine novel mutations in myosin VI, myosin VIIA and myosin XVA, doubling the number of myosin mutations in the Middle Eastern populations.
Two novel mutations were detected in the myosin VI, c.897G>T and p.L926Q, both in Palestinian Arab families (Table 1). The c.897G>T is a splice mutation, leading to the skipping of exon 10 (Figure 1c). This variant is located in the motor domain of the myosin VI protein in a region that interacts with a critical element of the active site called Switch I39 (Figure 1f). The mode of inheritance appears to be autosomal semi-dominant, as homozygotes for the mutation show a more severe phenotype with early onset, compared with the hearing-impaired heterozygotes. For c.897G>T, as exon 10 codes for an essential region of the motor, it may be that the protein by the mutant allele produced is non-functional and unstable, resulting in haploinsufficiency for myosin VI in these patients. The p.L926Q variant is located in a region rich in charged residues, which has been proposed to form a single alpha helix (SAH) and is also a potential proximal dimerization region, in the tail region of the myosin VI protein (http://www.uniprot.org/uniprot/)40 (Figure 1f). A mutation in this region, which has been proposed to form a SAH in the monomeric motor molecule, should not have a drastic effect on the monomeric motor. However, this region has also been predicted to be key for the proximal dimerization of the motor, suggesting that this mutation is likely to have an effect on stereociliar function that requires myosin VI to function as a dimer. These two mutations are the first mutations in MYO6 identified among the deaf population of the Middle East.
The five recessive novel mutations detected in MYO7A include three Israeli Jewish and two Palestinian Arab mutations. The three missense mutations are positioned in the motor domain of the myosin VIIA protein and were superimposed on a structural model of the myosin VIIa motor domain, based on the structure of myosin V bound to ATP (1W7j)38 (Figure 2i). Asparagine 207 is a critical residue of the active site essential for the binding of the nucleotide and its hydrolysis. The p.Asn207Ser mutation thus results in a motor unable to produce force that likely has a strong affinity for F-actin. Structural rearrangements in the motor domain must be coordinated between three different allosteric sites: the active site in which ATP binds and is hydrolyzed, the actin-binding interface and the lever arm that includes the converter, which rotates upon force production in order to amplify rearrangements of the motor domain and produce movement on actin. Arginine 657 corresponds to a well-conserved buried arginine close to the connectors that direct the conformational changes of the lever arm when the motor explores the different states of the active site along the motor cycle (Figure 2i). Its mutation to a tryptophan is predicted to impair the cyclic rearrangements of the motor and thus lead to loss of myosin VIIa activity. We predict that that this variant would be pathogenic, at least with regards to hearing loss. p.Arg657Trp was presented as an unclassified ‘probably neutral’ variant previously, without any further analysis.41 It is more difficult to predict how the p.Val10Ala mutation affects myosin VIIA, as the N-terminal region is not conserved with other myosins whose structure is already known. In fact, the mutation to alanine would not be predicted to have much of an effect unless it destabilizes the folding of this region. However, its pathogenic role is most likely related to its interaction with the p.Arg657Trp mutation, also present in a heterozygote state in the patient. In one case, patient D114 with the myosin VIIA p.Asn207Ser mutation was also found to be heterozygous for the PCDH15 p.Arg245* mutation. At age 40, she presented with non-syndromic congenital profound HL. Sanger sequencing of the complete coding region of PCDH15 revealed no additional mutations in this gene. Each of her three hearing siblings carried none or only one of these two mutations. As myosin VIIA and PCDH15 interact with each other and are involved in the same pathway, this might suggest digenic inheritance.42 Further evidence for this possibility has been demonstrated in mice with mutations in both Myo7a and Pcdh15, which had a more severe hearing loss than with each mutation alone, demonstrating digenic inheritance.43
The p.Gln2071* nonsense mutation, located in the FERM 2 domain of the tail of the myosin VIIA protein (Figure 2j), is predicted to create a stop codon at exon 45 (out of 49) and thus lead to a truncated protein. The myosin C-terminal tail confers the specificity of myosin VIIA by its binding to different partners, and this mutation might lead to an unstable protein, preventing the ligand from binding. The c.4153-2A>G putative splice mutation, detected in two Palestinian Arab families enrolled in the study, is located in the FERM 1 domain of the tail of the myosin VIIA protein (Figure 2j). This mutation is positioned two nucleotides upstream of exon 33 and abolishes the 3’ consensus AG splice-site sequence by converting it to GG. This cDNA region could not be amplified by PCR but analysis of the mutation by the ASSP tool for splice-site prediction demonstrates that no splice site exists at this position in the mutated DNA. Before this study, one MYO7A mutation was known in the Middle East population among the Palestinian Arabs for non-syndromic hearing loss.22 Previously, mutations for a syndromic form of deafness, Usher syndrome type IB, had been identified in this gene in Israel.44, 45 Targeted MPS enabled the discovery of an additional five novel mutations in this gene among both the Israeli Jewish and the Palestinian Arab populations, suggesting that more unidentified mutations may exist.
The novel missense mutations in myosin XVA include p.Ala408Val in compound heterozygosity with the known p.Glu1414Lys mutation in a Palestinian Arab family. There is uncertainty as to the deleterious nature of the p.Ala408Val mutation located at the N-terminal region before the motor domain of the myosin XVA protein, as the structure cannot be predicted for this region (Figure 3e). The bioinformatics analysis is inconsistent, though there are suggestions that this variant leads to changes in protein features and a splice site, potentially affecting protein function. Its interaction with the second mutation, p.Glu1414Lys, may be the key to its pathogenicity.
The myosin XVA p.Asp2823Asn novel mutation was found in two Israeli Jewish families originating from Iran, as well as in two out of 171 Jewish Iranian controls. It was not detected among any other Jewish ethnic groups, suggesting a founder effect in the Jewish Iranian population. This mutation was located in the tail domain of the myosin XVA protein (Figure 3e) in the F3 subdomain of the first FERM motif. The conservation with current structures of MythFERM domains is not high enough to predict more precisely what would be the impact of this mutation, namely, whether it affects a residue on the surface or whether this mutation would affect the stability of the domain. However, bioinformatics analysis with protein prediction programs demonstrates that this is a rather conservative one, suggesting that a change in this region would be pathogenic. These two novel mutations raise the number of MYO15A mutations in the Middle East to six (Table 1).
Seven out of the nine novel mutations identified in this study were not detected in additional families, emphasizing the known phenomenon of many deafness-causing mutations reported in single families worldwide. This further supports the conclusion that deep sequencing is the optimal genetic diagnostic approach for identification of mutations responsible for hearing loss. Myosin mutations span all regions of these molecular motors, with a wide range of hearing phenotypes, demonstrating the key role this family of proteins has on the function and well-being of the inner ear. Myosins may have an even larger impact on auditory function than in these isolated cases of monogenic deafness. Progressive hearing loss associated with MYO6 mutations resemble presbycusis,46 suggesting that mutations in myosins may be responsible for age-related hearing impairment. The discovery of these mutations will be facilitated by deep sequencing.
References
Van Camp G, Smith RJH Hereditary Hearing Loss Homepage. Available at http://hereditaryhearingloss.org. Accessed July 2013.
Shearer AE, DeLuca AP, Hildebrand MS et al: Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci USA 2010; 107: 21104–21109.
Brownstein Z, Friedman LM, Shahin H et al: Targeted genomic capture and massively parallel sequencing to identify genes for hereditary hearing loss in Middle Eastern families. Genome Biol 2011; 12: R89.
Odronitz F, Kollmar M : Drawing the tree of eukaryotic life based on the analysis of 2269 manually annotated myosins from 328 species. Genome Biol 2007; 8: R196.
Hartman MA, Spudich JA : The myosin superfamily at a glance. J Cell Sci 2012; 125: 1627–1632.
Petit C, Richardson GP : Linking genes underlying deafness to hair-bundle development and function. Nat Neurosci 2009; 12: 703–710.
Walsh T, Walsh V, Vreugde S et al: From flies' eyes to our ears: mutations in a human class III myosin cause progressive nonsyndromic hearing loss DFNB30. Proc Natl Acad Sci USA 2002; 99: 7518–7523.
Melchionda S, Ahituv N, Bisceglia L et al: MYO6, the human homologue of the gene responsible for deafness in Snell's waltzer mice, is mutated in autosomal dominant nonsyndromic hearing loss. Am J Hum Genet 2001; 69: 635–640.
Ahmed ZM, Morell RJ, Riazuddin S et al: Mutations of MYO6 are associated with recessive deafness, DFNB37. Am J Hum Genet 2003; 72: 1315–1322.
Liu XZ, Walsh J, Tamagawa Y et al: Autosomal dominant non-syndromic deafness caused by a mutation in the myosin VIIA gene. Nat Genet 1997; 17: 268–269.
Weil D, Kussel P, Blanchard S et al: The autosomal recessive isolated deafness, DFNB2, and the Usher 1B syndrome are allelic defects of the myosin-VIIA gene. Nat Genet 1997; 16: 191–193.
Wang A, Liang Y, Fridell RA et al: Association of unconventional myosin MYO15 mutations with human nonsyndromic deafness DFNB3. Science 1998; 280: 1447–1451.
Lalwani AK, Goldstein JA, Kelley MJ, Luxford W, Castelein CM, Mhatre AN : Human nonsyndromic hereditary deafness DFNA17 is due to a mutation in nonmuscle myosin. MYH9. Am J Hum Genet 2000; 67: 1121–1128.
Donaudy F, Snoeckx R, Pfister M et al: Nonmuscle myosin heavy-chain gene MYH14 is expressed in cochlea and mutated in patients affected by autosomal dominant hearing impairment (DFNA4). Am J Hum Genet 2004; 74: 770–776.
Donaudy F, Ferrara A, Esposito L et al: Multiple mutations of MYO1A, a cochlear-expressed gene, in sensorineural hearing loss. Am J Hum Genet 2003; 72: 1571–1577.
Zadro C, Alemanno MS, Bellacchio E et al: Are MYO1C and MYO1F associated with hearing loss? Biochim Biophys Acta 2009; 1792: 27–32.
Weil D, Blanchard S, Kaplan J et al: Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature 1995; 374: 60–61.
Mohiddin SA, Ahmed ZM, Griffith AJ et al: Novel association of hypertrophic cardiomyopathy, sensorineural deafness, and a mutation in unconventional myosin VI (MYO6). J Med Genet 2004; 41: 309–314.
Gibson F, Walsh J, Mburu P et al: A type VII myosin encoded by the mouse deafness gene shaker-1. Nature 1995; 374: 62–64.
Avraham KB, Hasson T, Steel KP et al: The mouse Snell's waltzer deafness gene encodes an unconventional myosin required for structural integrity of inner ear hair cells. Nat Genet 1995; 11: 369–375.
Probst FJ, Fridell RA, Raphael Y et al: Correction of deafness in shaker-2 mice by an unconventional myosin in a BAC transgene. Science 1998; 280: 1444–1447.
Shahin H, Walsh T, Rayyan AA et al: Five novel loci for inherited hearing loss mapped by SNP-based homozygosity profiles in Palestinian families. Eur J Hum Genet 2010; 18: 407–413.
Brownstein Z, Avraham KB : Deafness genes in Israel: implications for diagnostics in the clinic. Pediatr Res 2009; 66: 128–134.
Walsh T, Pierce SB, Lenz DR et al: Genomic duplication and overexpression of TJP2/ZO-2 leads to altered expression of apoptosis genes in progressive nonsyndromic hearing loss DFNA51. Am J Hum Genet 2010; 87: 101–109.
Edvardson S, Jalas C, Shaag A et al: A deleterious mutation in the LOXHD1 gene causes autosomal recessive hearing loss in Ashkenazi Jews. Am J Med Genet A 2011; 155A: 1170–1172.
Shahin H, Rahil M, Abu Rayan A et al: Nonsense mutation of the stereociliar membrane protein gene PTPRQ in human hearing loss DFNB84. J Med Genet 2010; 47: 643–645.
Walsh T, Shahin H, Elkan-Miller T et al: Whole exome sequencing and homozygosity mapping identify mutation in the cell polarity protein GPSM2 as the cause of nonsyndromic hearing loss DFNB82. Am J Hum Genet 2010; 87: 90–94.
Horn HF, Brownstein Z, Lenz DR et al: The LINC complex is essential for hearing. J Clin Invest 2013; 123: 740–750.
Parzefall T, Shivatzki S, Lenz DR et al: Cytoplasmic mislocalization of POU3F4 due to novel mutations leads to deafness in humans and mice. Hum Mut 2013; 34: 1102–1110.
Li H, Durbin R : Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010; 26: 589–595.
Albers CA, Lunter G, MacArthur DG, McVean G, Ouwehand WH, Durbin R : Dindel: accurate indel calls from short-read data. Genome Res 2011; 21: 961–973.
Chen K, Wallis JW, McLellan MD et al: BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods 2009; 6: 677–681.
Adzhubei IA, Schmidt S, Peshkin L et al: A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249.
Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC : SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 2012; 40: W452–W457.
Schwarz JM, Rodelsperger C, Schuelke M, Seelow D : MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010; 7: 575–576.
Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N : ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 2010; 38: W529–W533.
Coureux PD, Wells AL, Menetrey J et al: A structural state of the myosin V motor without bound nucleotide. Nature 2003; 425: 419–423.
Coureux PD, Sweeney HL, Houdusse A : Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J 2004; 23: 4527–4537.
Pylypenko O, Song L, Squires G et al: Role of insert-1 of myosin VI in modulating nucleotide affinity. J Biol Chem 2011; 286: 11716–11723.
Mukherjea M, Llinas P, Kim H et al: Myosin VI dimerization triggers an unfolding of a three-helix bundle in order to extend its reach. Mol Cell 2009; 35: 305–315.
Le Quesne Stabej P, Saihan Z, Rangesh N et al: Comprehensive sequence analysis of nine Usher syndrome genes in the UK National Collaborative Usher Study. J Med Genet 2012; 49: 27–36.
Senften M, Schwander M, Kazmierczak P et al: Physical and functional interaction between protocadherin 15 and myosin VIIa in mechanosensory hair cells. J Neurosci 2006; 26: 2060–2071.
Zheng QY, Scarborough JD, Zheng Y, Yu H, Choi D, Gillespie PG : Digenic inheritance of deafness caused by 8J allele of myosin-VIIA and mutations in other Usher I genes. Hum Mol Genet 2012; 21: 2588–2598.
Adato A, Weil D, Kalinski H et al: Mutation profile of all 49 exons of the human myosin VIIA gene, and haplotype analysis, in Usher 1B families from diverse origins. Am J Hum Genet 1997; 61: 813–821.
Rizel L, Safieh C, Shalev SA et al: Novel mutations of MYO7A and USH1G in Israeli Arab families with Usher syndrome type 1. Mol Vis 2011; 17: 3548–3555.
Oonk AM, Leijendeckers JM, Lammers EM et al: Progressive hereditary hearing impairment caused by a MYO6 mutation resembles presbyacusis. Hear Res 2013; 299: 88–98.
Acknowledgements
We thank the families for their kind participation in our research. We also thank Shaked Shivatzki for generating the figures. This work was supported by National Institutes of Health (NIDCD) R01DC011835 (KBA, MK), I-CORE Gene Regulation in Complex Human Disease Center no. 41/11 (KBA), Hedrich Charitable Trust (KBA), FRM équipes (AH), ANR blanche BLAN 2010 1504 01 (AH) and the ARC subvention fixe (SFI20121205398) (AH). This study has been performed by a team (AH) belonging to the Labex Deep: 11-LBX-0044.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Brownstein, Z., Abu-Rayyan, A., Karfunkel-Doron, D. et al. Novel myosin mutations for hereditary hearing loss revealed by targeted genomic capture and massively parallel sequencing. Eur J Hum Genet 22, 768–775 (2014). https://doi.org/10.1038/ejhg.2013.232
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2013.232
Keywords
This article is cited by
-
How myosin VI traps its off-state, is activated and dimerizes
Nature Communications (2023)
-
Analysis of the genotype–phenotype correlation of MYO15A variants in Chinese non-syndromic hearing loss patients
BMC Medical Genomics (2022)
-
A synonymous variant in MYO15A enriched in the Ashkenazi Jewish population causes autosomal recessive hearing loss due to abnormal splicing
European Journal of Human Genetics (2021)
-
Genotype-phenotype correlation analysis of MYO15A variants in autosomal recessive non-syndromic hearing loss
BMC Medical Genetics (2019)
-
Effects of genetic correction on the differentiation of hair cell-like cells from iPSCs with MYO15A mutation
Cell Death & Differentiation (2016)