Abstract
This study aimed to identify genetic mechanisms underlying severe retinal degeneration in one large family from northern Sweden, members of which presented with early-onset autosomal recessive retinitis pigmentosa and juvenile macular dystrophy. The clinical records of affected family members were analysed retrospectively and ophthalmological and electrophysiological examinations were performed in selected cases. Mutation screening was initially performed with microarrays, interrogating known mutations in the genes associated with recessive retinitis pigmentosa, Leber congenital amaurosis and Stargardt disease. Searching for homozygous regions with putative causative disease genes was done by high-density SNP-array genotyping, followed by segregation analysis of the family members. Two distinct phenotypes of retinal dystrophy, Leber congenital amaurosis and Stargardt disease were present in the family. In the family, four patients with Leber congenital amaurosis were homozygous for a novel c.2557C>T (p.Q853X) mutation in the CRB1 gene, while of two cases with Stargardt disease, one was homozygous for c.5461-10T>C in the ABCA4 gene and another was carrier of the same mutation and a novel ABCA4 mutation c.4773+3A>G. Sequence analysis of the entire ABCA4 gene in patients with Stargardt disease revealed complex alleles with additional sequence variants, which were evaluated by bioinformatics tools. In conclusion, presence of different genetic mechanisms resulting in variable phenotype within the family is not rare and can challenge molecular geneticists, ophthalmologists and genetic counsellors.
Similar content being viewed by others
Introduction
Leber congenital amaurosis (LCA) is a severe retinal dystrophy with onset of disease in early childhood. LCA is characterised by poor visual function, photophobia, high hyperopia, nystagmus, and severe retinal dysfunction.1 The electroretinogram is usually undetectable or severely reduced. Mode of inheritance for LCA is typically autosomal recessive and diagnosis of disease is established by clinical findings, although molecular genetic testing is available for 15 genes. The following genes are known to be associated with LCA: GUCY2D, RPE65, SPATA7, AIPL1, LCA5, RPGRIP1, CRX, CRB1, CEP290, IMPDH1, RD3, RDH12, KCNJ13, LRAT and TULP1.2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 It is difficult to estimate the proportion of patients with mutations in the different genes, as some, such as IMPDH1 (LCA11) is considered to be rare, other, as CEP290 accounts for almost 20%, and for some such as TULP1 and LRAT the number is uncertain. One of the most studied LCA genes is CRB1 at 1q31q32.1, which consists of 12 exons and encodes a protein Crumbs homologue that participates in determination and maintenance of photoreceptor architecture. Depending on the nature of the mutation, sequence changes in CRB1 can be disease causative in either LCA, retinitis pigmentosa (RP)18 or RP with preserved para-arteriolar retinal pigment epithelium.19
Stargardt disease (STGD1) is another autosomal recessive trait representing a severe form of retinal degeneration affecting the macula that begins in childhood. The gene responsible for STGD1 is ABCA4 at 1p22, which contains 50 exons and encodes a protein involved in energy transport to and from photoreceptor cells in the retina. Expressed exclusively in retinal photoreceptors, ABCA4 is involved in clearance of all-trans-retinal aldehyde that is a by-product of the retinoid cycle.20 Clinical diagnosis of the disease is difficult during eye examinations in the first few years of onset when discrete yellow spots or atrophy are occasionally seen in the macula. So far, more than 600 ABCA4 mutations scattered throughout the coding sequence have been annotated.21 In the same way as for the LCA-related genes, mutations in ABCA4 cause not only STGD1 but also cone-rod dystrophy and RP.22, 23
Molecular genetic testing is desirable for facilitating the diagnosis of LCA, early-onset RP and STGD1. This study was conducted to investigate the genetic defects in a Swedish family that manifests two distinct retinal degenerations: STGD1 and LCA.
Materials and methods
Patients and Clinical Examination
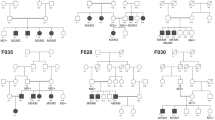
DNA was available from 6 affected individuals and 10 unaffected relatives from a multigeneration family originating from Jämtland County in northern Sweden (Figure 1a). In total, 356 control samples from a matched population were included in the study. Informed consent was obtained from all individuals participating in the study; the research followed the tenets of the Helsinki Declaration and was approved by the Ethics Committee of University of Umeå.
Pedigree of the Swedish family segregating CRB1 and ABCA4 mutations. (a) DNA from index patient VI:2 (marked by an arrow) and V:4 were used for targeted mutation screening (APEX). Affected individuals are shown in shaded red or blue and healthy subjects are shown as open circles for females and squares for males. CRB1 and ABCA4 mutant alleles corresponding LCA and STGD1 phenotypes are drawn in red and blue, respectively. * denotes that DNA was not available. (b) Segregation analysis of a novel CRB1 c.2557C>T (p.Q853X) mutation in the Swedish family was carried out by PCR-restriction fragment length polymorphism analysis of the CRB1 exon 7. The 472-bp-long PCR products amplified according to the conditions described in ‘Materials and Methods’ were digested with DdeI endonuclease and results were visualised on 2% agarose gel (SeaKem, ME Agarose, Lonza, Basel, Switzeland) by GelStar Nuclear Acid Staining (Cambrex, Bio Science Rockland Inc, Rockland, ME, USA). c.2557C>T mutation abolishes DdeI restriction site. LCA cases are indicated in red and STGD1 in blue.
Clinical ophthalmological and electrophysiological examinations were performed and previous examination results of affected individuals along with their family history were collected from their home clinics. Full-field, single-flash, flicker electroretinograms and oscillatory potentials were recorded (UTAS-E 2000, LKC Technologies Inc., Gaithersburg, MD, USA) according to the recommendations of the International Society for Clinical Electrophysiology of Vision.24
Molecular genetic analysis
DNA from 16 individuals was extracted from peripheral blood lymphocytes using a previously described protocol.25 Testing for the mutations reported as a cause of LCA, autosomal recessive RP and Stargardt disease was performed by arrayed primer extension (APEX) at AsperBiotech (Tartu, Estonia) (http://www.asperbio.com/asper-ophthalmics). High-resolution genome-wide SNP-array genotyping was applied for identification of homozygosity regions containing potential disease-causing genes. DNA was genotyped on a SNP microarray (Human610-Quad BeadChip, Illumina, San Diego, CA, USA) with 610 000 polymorphic SNPs according to manufacturer’s instructions. The data were analysed using GenomeStudio software (Illumina). Genomic regions demonstrating homozygosity over 5 Mb were taken into account for further gene analysis.
For bi-directional sequencing of CRX1 (MIM 602225, NM_000554.4), CRB1 (MIM 604210, NM_201253.2) and ABCA4 (MIM 601691, NM_000350.2), coding exons and adjacent intronic sequences were amplified from genomic DNA. Primer pairs for CRX1 and CRB1 were designed with Primer3 software and are available upon request. PCR amplification and the sequencing reactions were performed as described elsewhere.26 The products of sequencing reactions were analysed on ABI 3500xL Dx Genetic Analyser (Applied Biosystems, Foster City, CA, USA). Sequences were aligned and evaluated using the Sequencher software version 4.9 (Gene Codes Corporation, Ann Arbor, MI, USA). Sequencing of the entire coding region of ABCA4 was performed at AsperBiotech (http://www.asperbio.com/genetic-tests/panel-of-genetic-tests/stargardt-disease-cone-rod-dystrophy-abca4).
All identified variants were denoted using accepted nomenclature recommended by Human Genome Variation Society. To predict the impact of sequence variants on the CRB1 and ABCA4 function, missense mutations were analysed by Sorting Intolerant from Tolerant (SIFT; http://sift.jcvi.org) and Polymorphism Phenotyping (PolyPhen; http://genetics.bwh.harvard.edu/pph). Variants detected in intronic sequences were analysed with the splice site prediction programs GeneSplicer (http://www.cbcb.umd.edu/software/GeneSplicer) and Splice Site Finder (www.genet.sickkids.on.ca/_ali/splicesitefinder). All bioinformatics tools were available via the Alamut software version 2.0 (Interactive Biosoftware, Rouen, France).
Results
Clinical findings
In total, six affected patients with retinal degeneration were identified in a large Swedish family (Figure 1a). Two different phenotypes were recognised; four of the cases (Figure 1a, VI:2, VI:5, VI:6, VII:3) presented an early-onset RP, LCA, and two of the cases (Figure 1a, V:4, VI:10) presented an early-onset maculopathy, Stargardt disease (STGD1).
The LCA cases showed severely reduced visual acuity or blindness in childhood, as well as nystagmus, convergent strabismus and severe hyperopia. In early childhood, the macular area showed central diffuse atrophy and the peripheral retina had an overall granulated appearance. In the late thirties, premature cataract evolved and synchysis scintillans were observed in the vitreous body in 2/4 LCA cases. In the central retina, areolar atrophy surrounded by prominent pigmentation of the macular area was found with generally dispersed pigmentary changes of more peripheral retina (Figure 2a). No preservation of the para-arteriolar pigment epithelium or peripheral telangiectasia was observed in the LCA cases in adulthood. The full-field electroretinogram responses described at the age of 3 years were extinguished in early childhood.
Fundus appearance in patients with CRB1 and ABCA4 mutations. (a) LCA patient, VI:2, 43-year-old woman with LCA. Peripheral pigmentary changes with extensive macular atrophy and drusen of the optic disc are shown. (b) STGD1 patient,V:4, 56-year-old man. An extensive macular atrophy with pigmentation is shown; the atrophy is extending into the peripheral parts of the retina. (c) STGD1 patient 41, VI:10, 25-year-old woman. Central retinal atrophy of the macula with hyperpigmentations and dispersed yellowish flecks of the posterior pole are visible.
The STGD1 cases demonstrated a different retinal phenotype. Visual acuity was affected at school age (8–14 years). Macular atrophy was present with some hyperpigmentation and yellowish flecks of the posterior pole with central visual field defects (Figures 2b and c). The peripheral retina was preserved in young adulthood, although progressive retinal atrophy, peripheral retinal function and visual fields diminished in adulthood. No premature cataract was observed. In adulthood (VI:10, age 25 years), the recovery of standardised dark adaptation showed both cone and rod adaptation, with a final visual sensory threshold elevated by 1 log unit. In the full-field electroretinograms, the rod, mixed rod–cone and cone amplitudes were estimated within normal range but the 30-Hz flicker amplitude was decreased to 40% of normal level with prolonged implicit time.
Molecular genetic findings
Genetic testing for known mutations
DNA from two patients, one with LCA (VI:2) and another one with STGD1 (V:4), were analysed for the presence of previously reported sequence variants known as a cause or associated with autosomal recessive RP, LCA and STGD1. In total, 641 mutations in 13 LCA genes and 594 mutations in 19 RP genes on genotyping microarrays (commercially designed gene chips available via http://www.asperbio.com/asper-ophthalmics) were tested by APEX. In LCA patient VI:2, no mutations were identified by APEX analysis. This patient was heterozygous carrier of four SNPs, all predicted to be tolerated according to SIFT and PolyPhen (Table 1). In STGD1 patient V:4, only one heterozygous variant c.5461-10T>C in the ABCA4 gene was detected, which was predicted to have a weak splice effect.
Homozygosity regions detection
Absence of detected mutations in the LCA patient, recessive inheritance pattern and presence of consanguinity loops in the family (Figure 1a) encouraged us to undertake high-resolution SNP-array genotyping on both patients aiming to reveal regions of homozygosity containing potential disease-causing genes. In STGD1 patient V:4, only one 2.2 Mb homozygous region was identified that did not contain any known retinal genes and did not overlap with any of eight regions of homozygosity (ROH) detected in LCA individual VI:2 (Table 2). Two of the eight ROH on chromosomes 1 and 19 contained genes responsible for retinal degeneration. These genes, OPA3, causing autosomal recessive optic atrophy with chorea and spastic paraplegia, PRPF31, causing an autosomal dominant form of RP with reduced penetrance and CFH, increasing the likelihood of developing age-related macular degeneration, were excluded as a potential cause of the disease in this family due to inheritance mode and absence of specific clinical appearance. Of the remaining candidate genes, the cone-rod homeobox gene (CRX1) on chromosome 19, known as a very rare cause of both dominant and recessive LCA, was less prioritised than the Crumbs homologue gene (CRB1) on chromosome 1, which was the most promising candidate gene responsible for the LCA phenotype in the family.
Sequence analysis of CRX1 and CRB1
Sequencing of the CRX1 in LCA patient VI:2 did not reveal any mutations and therefore this gene was excluded as a cause of the disease in the family. Sequencing of all 12 exons of CRB1 in the same patient identified a homozygous nonsense mutation c.2557C>T in exon 7, resulting in a premature stop codon and a truncated protein, p.Q853X (Figure 3a). Segregation analysis of affected and unaffected members of the family was performed by restriction fragment length polymorphism analysis, using the DdeI endonuclease for digestion of CRB1 exon 7. The c.2557C>T (p.Q853X) mutation was present in homozygous form in all four LCA patients (Figure 1a and b, VI:2, VI:5, VI:6, VII:3), and in heterozygous form in the healthy parents of individual VII:3. It was absent in 356 control samples from a matched population. The c.2557C>T (p.Q853X) mutation has not been described in the literature previously and is thus novel to this Swedish family.
CRB1 and ABCA4 mutations causing LCA and STGD1 in the Swedish family. Sequence analysis demonstrating sequence variants CRB1 c.2557C>T (a), reverse sequence), ABCA4 c.5461-10T>C (b), reverse sequence) and ABCA4 c.4773+3A>G (c), forward sequence). The upper images show wild-type sequences, the middle images show heterozygous mutations and low images show homozygous mutations. Mutations positions are marked in black.
Sequence analysis of ABCA4
Sequence analysis of ABCA4 intron 38 in STGD1 patient V:4 confirmed heterozygosity of the mutation c.5461-10T>C detected in the APEX analysis (Figure 3b). To determine if individual V:4 was a compound heterozygote, all exons and flanking intronic sequences of the ABCA4 were sequenced. In total, 12 sequence variants were detected, 8 of which were intronic and 4 exonic (Table 3). Of the intronic variants, the most interesting was a novel sequence variant in ABCA4 intron 33, c.4773+3A>G (Figure 3c), which was predicted to have a weak splice effect. Of the exonic variants, only p.N1868I and p.H423R were non-synonymous. Bioinformatics analysis predicted p.N1868I to be possibly damaging for protein function, whereas p.H423R was predicted to be benign.
Segregation analysis was done for three ABCA4 sequence variants, including p.N1868I, c.4773+3A>G and c.5461-10T>C. The variant p.N1868I was found in homozygous form in STGD1 patient VI:10, and in heterozygous form in STGD1 patient V:4 and non-affected individuals V:6, V:7 and VI:8 (Figure 1a). To determine if p.N1868I was a common variant in our population, we tested 115 control individuals from a matched geographic region and detected 16 heterozygous carriers. This yielded an estimated allele frequency of 0.139, which is higher than the reported allele frequency in the SNP database (MAF=0.029).
The novel variant c.4773+3A>G was found in heterozygous form in the two LCA patients VI:2 and VI:5, in three unaffected individuals (V:5, VI:8, VI:9) and in STGD1 patient V:4 (Figure 1a). Testing of 113 matched healthy controls revealed one heterozygous carrier, yielding an allele frequency of 0.009.
The variant c.5461-10T>C was found in homozygous form in individual VI:10 (Figure 3b), and in heterozygous form in the healthy parents of this individual (V:6, V:7), as well as in STGD1 patient V:4. Testing of 116 clinically matched control individuals revealed no other carriers. As follows from segregation and haplotype analyses, STGD1 patient V:4 is presumably compound heterozygous for the two rare splice variants c.5461-10T>C and c.4773+3A>G.
Discussion
In this study, we approached patients with different clinical presentation belonging to the same multigeneration family of Swedish origin. Clinical diagnosis of LCA was recognised in four of six patients. Molecular testing of LCA patients is quite laborious due to genetic heterogeneity involving 16 known LCA genes. Testing for known mutations by array technology provides fast and reliable results; however, it does not reveal novel mutations. Therefore, arrays interrogating disease-causing genes need to be updated along with the discovery of novel mutations. Despite some disadvantages, in many cases this method is used as a first-line screening to reveal genetic mechanisms underlying different ophthalmic disorders. In our study, however, none of the known LCA mutations was identified in the index patient with LCA (VI:2), and only one heterozygous mutation in the ABCA4 gene was found in the STGD1 patient (V:4).
Recently, it has been shown that genome-wide homozygosity mapping with SNP microarrays represents a powerful tool for mutation discovery in autosomal recessive disorders such as LCA and RP.27, 28, 29 In one of these studies, identification of homozygosity regions and further sequencing of LCA genes within these regions resulted in detection of 10 homozygous mutations, 7 of which were novel.27 LCA genes CEP290 and LCA5 were identified by the same approach.7, 27 As inheritance pattern of the disease in our consanguineous family was autosomal recessive, we performed genome-wide genotyping in two affected patients, aiming at identification of homozygous regions. The LCA patient VI:2 demonstrated ROH of more than 5 Mb in size on six chromosomes, of which the most promising candidate gene was CRB1 on chromosome 1. Subsequent sequencing of CRB1 resulted in detection of a novel null mutation in exon 7, c.2557C>T (p.Q853X). Among 150 known CRB1 sequence variants, mutations in exon 7 are the most frequent (27%) and especially important for CRB1 function due to encoded laminin AG-like domain.30, 31
The c.5461-10T>C mutation was first reported by Maugeri et al.,32 although its function is still not resolved. The functional consequence of c.5461-10T>C was accessed in a study with the Exon Trapping System by Rivera et al.,33 who classified this nucleotide change as a rare sequence variant, as only correctly spliced exon was detected. The majority of STGD1 patients are compound heterozygotes, with our patient V:4 not being an exception, representing one of multiple cases with the c.5461-10T>C mutation. The c.5461-10T>C variant was found to be the most prevalent allele among patients with autosomal recessive cone and cone-rod dystrophy (8 of 64 patients).34 In another study, the variant was found in 27 of 518 STGD1 patients compared with 1 of 316 clinically matched control individuals.33 It is to be noticed that another affected member (VI:10) in the same family was homozygous for the c.5461-10T>C mutation. None of our healthy controls carried the c.5461-10T>C mutation, in line with previous observations of different population carrier frequencies.35
Interestingly, our STGD1 patients carried the sequence variant ABCA4 p.N1868I that was predicted to be possibly damaging, as well as acting as a risk-increasing factor in AMD.36 In our study, this variant was detected in almost 14% of the healthy controls, which is higher compared with the maximal frequency of 7.5% reported in a Finnish population in the 1000 Genomes project.36 It is worth mentioning that ABCA4 c.2588G>C (p.G863A), the most frequent autosomal recessive mutation in the European population, is disease causative only in combination with a severe ABCA4 mutation,32 and does not result in a STGD1 phenotype when present bi-allelic or in combination with a mild ABCA4 mutation. Maugeri et al.32 hypothesised the possibility of a carrier advantage due to the high carrier frequency of the 2588C allele in Sweden (1 out of 18),37 although the incidence of STGD1 is not higher in our country than in the rest of Europe. The same phenomena can also be applied to p.N1868I.
In the heterozygous STGD1 patient (V:4), we discovered a novel sequence variant c.4773+3A>G, which was predicted to reduce the strength of the donor splice site. While the manuscript was in preparation, this variant was also reported in one AMD patient.36 This variant was not detected in 3510 controls of European American descent,36 and in our study only one carrier of 113 tested was found. The effect of the c.4773+3A>G mutation can be tested using RNA splicing analysis. However, when we performed RT-PCR analysis on RNA isolated from peripheral blood lymphocytes from the heterozygous patient VI:2, only correctly spliced exon was detected (data not shown). This result was not unexpected, as ABCA4 mRNA is expressed exclusively in retina.38
In conclusion, we showed that in a large Swedish family, defects in two genes, CRB1 (c.2557C>T) and ABCA4 (c.5461-10T>C and c.4773+3A>G), caused two different retinal diseases. This is a second report presenting data on the mutations affecting CRB1 and ABCA4 genes segregating with two different phenotypes, namely LCA and autosomal recessive retinitis pigmntosa in the same family.39 Presence of different genetic mechanisms resulting in variable phenotype within the family probably is not such a rare event and should be considered in patient management and disease treatment.
References
Chung DC, Traboulsi EI : Leber congenital amaurosis: clinical correlations with genotypes, gene therapy trials update, and future directions. J AAPOS 2009; 13: 587–592.
Hanein S, Perrault I, Gerber S et al: Leber congenital amaurosis: comprehensive survey of the genetic heterogeneity, refinement of the clinical definition, and genotype-phenotype correlations as a strategy for molecular diagnosis. Hum Mutat 2004; 23: 306–317.
Senechal A, Humbert G, Surget MO et al: Screening genes of the retinoid metabolism: novel LRAT mutation in leber congenital amaurosis. Am J Ophthalmol 2006; 142: 702–704.
Sergouniotis PI, Davidson AE, Mackay DS et al: Recessive mutations in KCNJ13, encoding an inwardly rectifying potassium channel subunit, cause leber congenital amaurosis. Am J Hum Genet 2011; 89: 183–190.
Friedman JS, Chang B, Kannabiran C et al: Premature truncation of a novel protein, RD3, exhibiting subnuclear localization is associated with retinal degeneration. Am J Hum Genet 2006; 79: 1059–1070.
Bowne SJ, Sullivan LS, Mortimer SE et al: Spectrum and frequency of mutations in IMPDH1 associated with autosomal dominant retinitis pigmentosa and leber congenital amaurosis. Invest Ophthalmol Vis Sci 2006; 47: 34–42.
den Hollander AI, Koenekoop RK, Yzer S et al: Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet 2006; 79: 556–561.
den Hollander AI, Davis J, van der Velde-Visser SD et al: CRB1 mutation spectrum in inherited retinal dystrophies. Hum Mutat 2004; 24: 355–369.
Freund CL, Wang QL, Chen S et al: De novo mutations in the CRX homeobox gene associated with Leber congenital amaurosis. Nat Genet 1998; 18: 311–312.
Dryja TP, Adams SM, Grimsby JL et al: Null RPGRIP1 alleles in patients with Leber congenital amaurosis. Am J Hum Genet 2001; 68: 1295–1298.
den Hollander AI, Koenekoop RK, Mohamed MD et al: Mutations in LCA5, encoding the ciliary protein lebercilin, cause Leber congenital amaurosis. Nat Genet 2007; 39: 889–895.
Perrault I, Rozet JM, Calvas P et al: Retinal-specific guanylate cyclase gene mutations in Leber’s congenital amaurosis. Nat Genet 1996; 14: 461–464.
Marlhens F, Bareil C, Griffoin JM et al: Mutations in RPE65 cause Leber's congenital amaurosis. Nat Genet 1997; 17: 139–141.
Wang H, den Hollander AI, Moayedi Y et al: Mutations in SPATA7 cause Leber congenital amaurosis and juvenile retinitis pigmentosa. Am J Hum Genet 2009; 84: 380–387.
Sohocki MM, Bowne SJ, Sullivan LS et al: Mutations in a new photoreceptor-pineal gene on 17p cause Leber congenital amaurosis. Nat Genet 2000; 24: 79–83.
Janecke AR, Thompson DA, Utermann G et al: Mutations in RDH12 encoding a photoreceptor cell retinol dehydrogenase cause childhood-onset severe retinal dystrophy. Nat Genet 2004; 36: 850–854.
Berger W, Kloeckener-Gruissem B, Neidhardt J : The molecular basis of human retinal and vitreoretinal diseases. Prog Retin Eye Res 2010; 29: 335–375.
den Hollander AI, Heckenlively JR, van den Born LI et al: Leber congenital amaurosis and retinitis pigmentosa with Coats-like exudative vasculopathy are associated with mutations in the crumbs homologue 1 (CRB1) gene. Am J Hum Genet 2001; 69: 198–203.
den Hollander AI, ten Brink JB, de Kok YJ et al: Mutations in a human homologue of Drosophila crumbs cause retinitis pigmentosa (RP12). Nat Genet 1999; 23: 217–221.
Cideciyan AV, Aleman TS, Swider M et al: Mutations in ABCA4 result in accumulation of lipofuscin before slowing of the retinoid cycle: a reappraisal of the human disease sequence. Hum Mol Genet 2004; 13: 525–534.
Zernant J, Schubert C, Im KM et al: Analysis of the ABCA4 gene by next-generation sequencing. Invest Ophthalmol Vis Sci 2011; 52: 8479–8487.
Cremers FP, van de Pol DJ, van Driel M et al: Autosomal recessive retinitis pigmentosa and cone-rod dystrophy caused by splice site mutations in the Stargardt's disease gene ABCR. Hum Mol Genet 1998; 7: 355–362.
Maugeri A, Klevering BJ, Rohrschneider K et al: Mutations in the ABCA4 (ABCR) gene are the major cause of autosomal recessive cone-rod dystrophy. Am J Hum Genet 2000; 67: 960–966.
Marmor MF, Holder GE, Seeliger MW, Yamamoto S : Standard for clinical electroretinography (2004 update). Doc Ophthalmol 2004; 107–114.
Balciuniene J, Johansson K, Sandgren O, Wachtmeister L, Holmgren G, Forsman K : A gene for autosomal dominant progressive cone dystrophy (CORD5) maps to chromosome 17p12-p13. Genomics 1995; 30: 281–286.
Kohn L, Kadzhaev K, Burstedt MS et al: Mutation in the PYK2-binding domain of PITPNM3 causes autosomal dominant cone dystrophy (CORD5) in two Swedish families. Eur J Hum Genet 2007; 15: 664–671.
den Hollander AI, Lopez I, Yzer S et al: Identification of novel mutations in patients with Leber congenital amaurosis and juvenile RP by genome-wide homozygosity mapping with SNP microarrays. Invest Ophthalmol Vis Sci 2007; 48: 5690–5698.
Collin RW, van den Born LI, Klevering BJ et al: High-resolution homozygosity mapping is a powerful tool to detect novel mutations causative of autosomal recessive RP in the Dutch population. Invest Ophthalmol Vis Sci 2011; 52: 2227–2239.
Siemiatkowska AM, Arimadyo K, Moruz LM et al: Molecular genetic analysis of retinitis pigmentosa in Indonesia using genome-wide homozygosity mapping. Mol Vis 2011; 17: 3013–3024.
Benayoun L, Spiegel R, Auslender N et al: Genetic heterogeneity in two consanguineous families segregating early onset retinal degeneration: the pitfalls of homozygosity mapping. Am J Med Genet A 2009; 149A: 650–656.
Bujakowska K, Audo I, Mohand-Said S et al: CRB1 mutations in inherited retinal dystrophies. Hum Mutat 2012; 33: 306–315.
Maugeri A, van Driel MA, van de Pol DJ et al: The 2588G—>C mutation in the ABCR gene is a mild frequent founder mutation in the Western European population and allows the classification of ABCR mutations in patients with Stargardt disease. Am J Hum Genet 1999; 64: 1024–1035.
Rivera A, White K, Stohr H et al: A comprehensive survey of sequence variation in the ABCA4 (ABCR) gene in Stargardt disease and age-related macular degeneration. Am J Hum Genet 2000; 67: 800–813.
Kitiratschky VB, Grau T, Bernd A et al: ABCA4 gene analysis in patients with autosomal recessive cone and cone rod dystrophies. Eur J Hum Genet 2008; 16: 812–819.
Roberts LJ, Nossek CA, Greenberg LJ, Ramesar RS : Stargardt macular dystrophy: common ABCA4 mutations in South Africa—establishment of a rapid genetic test and relating risk to patients. Mol Vis 2012; 18: 280–289.
Fritsche LG, Fleckenstein M, Fiebig BS et al: A subgroup of age-related macular degeneration is associated with mono-allelic sequence variants in the ABCA4 gene. Invest Ophthalmol Vis Sci 2012; 53: 2112–2118.
Maugeri A, Flothmann K, Hemmrich N et al: The ABCA4 2588G>C Stargardt mutation: single origin and increasing frequency from South-West to North-East Europe. Eur J Hum Genet 2002; 10: 197–203.
Allikmets R : A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet 1997; 17: 122.
Riveiro-Alvarez R, Vallespin E, Wilke R et al: Molecular analysis of ABCA4 and CRB1 genes in a Spanish family segregating both Stargardt disease and autosomal recessive retinitis pigmentosa. Mol Vis 2008; 14: 262–267.
Acknowledgements
We thank all members of the family and acknowledge collaboration with AsperBiotech, Tartu, Estonia. We also thank Professor Anneke den Hollander for the discussion in the beginning of the study. This study was supported by grants from Visare Norr, Stiftelsen Kronprinsessan Margaretas Arbetsnämnd för synskadade and University Hospital of Umeå.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Jonsson, F., Burstedt, M., Sandgren, O. et al. Novel mutations in CRB1 and ABCA4 genes cause Leber congenital amaurosis and Stargardt disease in a Swedish family. Eur J Hum Genet 21, 1266–1271 (2013). https://doi.org/10.1038/ejhg.2013.23
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2013.23