Abstract
Charcot–Marie–Tooth disease (CMT) is a group of hereditary peripheral neuropathies. The dominantly inherited axonal CMT2 displays striking genetic heterogeneity, with 17 presently known disease genes. The large number of candidate genes, combined with lack of genotype–phenotype correlations, has made genetic diagnosis in CMT2 time-consuming and costly. In Finland, 25% of dominant CMT2 is explained by either a GDAP1 founder mutation or private MFN2 mutations but the rest of the families have remained without molecular diagnosis. Whole-exome and genome sequencing are powerful techniques to find disease mutations for CMT patients but they require large amounts of sequencing to confidently exclude heterozygous variants in all candidate genes, and they generate a vast amount of irrelevant data for diagnostic needs. Here we tested a targeted next-generation sequencing approach to screen the CMT2 genes. In total, 15 unrelated patients from dominant CMT2 families from Finland, in whom MFN2 and GDAP1 mutations had been excluded, participated in the study. The targeted approach produced sufficient sequence coverage for 95% of the 309 targeted exons, the rest we excluded by Sanger sequencing. Unexpectedly, the screen revealed a disease mutation only in one family, in the HSPB1 gene. Thus, new disease genes underlie CMT2 in the remaining families, indicating further genetic heterogeneity. We conclude that targeted next-generation sequencing is an efficient tool for genetic screening in CMT2 that also aids in the selection of patients for genome-wide approaches.
Similar content being viewed by others
Introduction
Charcot–Marie–Tooth disease (CMT) is an inherited disease of the peripheral nervous system with a prevalence of ∼1:2500, which makes it the most common genetic neuromuscular disorder.1 The pathogenic mechanism is a defect of myelination (CMT1) or axon function (CMT2). The genetic heterogeneity of CMT reflects the unique anatomy and metabolism of the peripheral nervous system. The number of CMT disease genes has grown rapidly since the first causative gene defect was identified in 1991.2 Defects in more than 40 genes are now known to be associated with CMT or related phenotypes, inherited through all possible Mendelian inheritance patterns, according to the Inherited Peripheral Neuropathy Mutation Database (IPNMD, http://www.molgen.ua.ac.be/CMTmutations/Mutations/).
CMT1 is typically caused by defects in genes expressed in Schwann cells, whereas CMT2 disease genes encode a functionally broader group of proteins required for axon maintenance.3 The molecular diagnosis of CMT1 is often straightforward as 70–80% of patients have a duplication or mutation in the gene encoding the peripheral myelin protein-22 (PMP22). Diagnosis of CMT2 relies on electrophysiologic studies, which typically demonstrate normal or slightly decreased nerve conduction velocity with decreased amplitude. At least 17 disease genes for CMT2 have been reported, complicating the molecular diagnosis in patients. Although recessive inheritance and de novo mutations are possible, most CMT2 cases follow autosomal dominant (AD) inheritance. Mutations in MFN2, encoding mitofusin2, have been reported in up to 20% of families with AD-CMT2.4 Mitofusins regulate mitochondrial fusion, thus being essential for mitochondrial dynamics in axons. Mutations in the other CMT2 genes explain the disease only in small percentages of patients. Of these genes, GDAP1 also encodes a protein involved in mitochondrial dynamics, whereas others encode components of axonal cytoskeleton (NEFL), proteins that regulate vesicular and mitochondrial transport along axons (KIF1B, DNM2, DYNC1H1, RAB7A, LRSAM), ubiquitous proteins involved in gene expression and protein synthesis (MED25, LMNA, AARS, GARS), calcium channels (TRPV4) or chaperones (HSPB1, HSPB8). The large number of potential disease genes has prevented simple molecular genetic diagnosis in most patients with CMT2.
Founder effects, mutation hot spots and chance influence the contribution of each gene to CMT2 in different populations. In our cohort of Finnish patients with dominant CMT2, a founder mutation in GDAP1 (leading to amino acid change H123R) explains 14% and private MFN2 mutations 11% of the familiar disease.5 The rest of the Finnish CMT2 patients lack molecular diagnosis because the screening of all potential candidate genes by Sanger sequencing has been too large of an undertaking for the health-care system. Therefore, a means to screen all candidate genes in a fast and cost-effective manner would be a major improvement.
Next-generation sequencing (NGS) methods are expected to dramatically advance the gene diagnosis process in CMT patients. Both whole-genome sequencing and whole-exome sequencing (WES) have already been successfully employed in CMT mutation discovery.6, 7, 8 One of the disadvantages of these methods is that they require very large amounts of sequencing to achieve sufficient read coverage for confidential exclusion of heterozygous variants in all candidate genes. For example in WES, the sequence reads are often unevenly distributed over the exome, and thus the average read coverage reveals little about the method’s power to find or exclude all possible disease-causing mutations. Through these methods a vast amount of unnecessary data are produced when the purpose in a diagnostic setting is only to analyze mutations in a determined set of candidate genes.6, 7, 8 We chose to test a targeted NGS approach for CMT2 gene diagnostics, focusing on confident exclusion of known disease genes.
Materials and methods
Patients
In total 15 unrelated patients of Finnish origin, diagnosed with familiar adult-onset CMT2, were included in the study. Electroneuromyography studies were performed on all patients. The findings were consistent with length-dependent sensory-motor and axonal neuropathy with normal (>38 m/s) or slightly decreased (25–38 m/s) nerve conduction velocities (NCV) typical for CMT2. Autosomal dominant inheritance pattern was suspected in all families. The clinical and electrophysiological studies were performed at the Helsinki University Central Hospital or Oulu University Hospital. All subjects gave written informed consent and the study was approved by the ethics board of the Helsinki University Central Hospital.
Targeted NGS
The sequencing target was designed with the SureDesign program (Agilent Technologies, Santa Clara, CA, USA) to cover 17 CMT2 disease genes and an additional 143 candidate genes. Target enrichment and amplification was done with the HaloPlex Target Enrichment Kit (Agilent Technologies), according to the manufacturer’s instructions. The genomic patient DNA was first fragmented with restriction enzymes followed by hybridization of the target DNA to a biotinylated probe library and by its capture using streptavidin-coated magnetic beads. Finally, the target was PCR amplified to produce a target-enriched sample for sequencing. Sequencing was done on an MiSeq sequencer (Illumina, San Diego, CA, USA) and the reads were aligned to the target. The variant calling pipeline developed by the Finnish Institute of Molecular Medicine9 was used for variant calling. The CMT2 disease gene exons that contained at least one nucleotide with a sequencing read depth of <6 were defined as not adequately covered by the NGS method. These exons were selected for Sanger sequencing. Primer sequences are listed in the Supplementary Table 3. Identified disease-causing variants were submitted to the ClinVar database (www.ncbi.nlm.nih.gov/clinvar/).
Results
Patients
In total, 15 unrelated patients were included in the study with inclusion criteria of a diagnosis of axonal CMT at age 17 or older by clinical assessment and electrophysiological study, positive family history with suspicion of dominant inheritance, and consent to enroll in the study. The patients were prescreened for the GDAP1 c.368 A>G (p.(His123Arg)) (RefSeq NM_018972.2) mutation and MFN2 coding-region mutations, with negative results.
Targeted NGS in CMT2 patients
We selected the 309 coding exons of 17 previously reported CMT2 disease genes (KIF1B, MFN2, RAB7A, TRPV4, GARS, NEFL, HSPB1, HSPB8, AARS, DNM2, DYNC1H1, MPZ, LRSAM1, GDAP1, MED25, LMNA, and BSCL2 (MIM#118210 entry, last updated 10/10/2012)) as the target for the NGS. For target enrichment we used the HaloPlex Target Enrichment System, which utilizes restriction enzymes to fragment genomic DNA. As this system allowed simultaneous analysis of up to 500 kb of target, we included an additional 143 candidate genes to a total target of 479 992 bp (Supplementary Table 1). These additional genes were selected primarily based on association with other neuropathies or due to a functional relation to a known CMT2 disease gene. Additionally, we included a number of genes in which candidate mutations had been previously identified by exome sequencing of CMT2 patients.5 The target translated into a total bait of 1.18 Mb, which included intronic flanking regions for each exon. To achieve sufficient target coverage, a minimum sequence of 100 Mb was generated. This requirement was not met for two of our samples, which resulted in their exclusion from full analysis. For the remaining 13 samples, mean bait coverage was 108X.
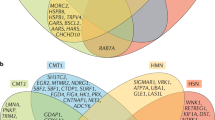
To evaluate the completeness of the method for the screening of the 17 known disease genes, we analyzed the read coverage of each coding exon of these genes in detail using the Integrative Genomics Viewer (Broad Institute). A minimum read coverage of six was regarded as necessary for identification or exclusion of a mutation. This was achieved for 293 of the 309 target exons (94.8%). The 16 remaining exons contained at least one insufficiently covered base pair, which was analyzed to result from the lack of suitable restriction enzyme recognition sites in the intended region. These sequences had thus not been included in the bait, indicating that additional rounds of sequencing would not have improved their coverage. The nine genes that were fully covered contained between 3 and 19 exons (Figure 1). The missed exons were distributed randomly within the remaining eight genes (1 per 20 exons on average), with the exception of GARS, which had as many as 5 exons out of 16 missing coverage for at least one base pair (Figure 1). To fully exclude all the known CMT2 genes, we designed primers for these 16 remaining exons and Sanger sequenced them.
Targeted NGS data reveal a new HSPB1 mutation in a large family with AD-CMT2
To find potential disease-causing mutations in the 17 known CMT2 disease genes, the NGS data were filtered through the following steps: (1) exclusion of variants with >0.005 frequency in the 1000Genomes database (www.1000genomes.org), (2) exclusion of variants that were not in the coding regions or in splice sites, (3) exclusion of synonymous variants. These steps resulted in only four remaining variants in the datasets of the 13 fully analyzed patients (Figure 2). The variants c.1280T>C (p.(Leu427Pro)) in BSCL2 (RefSeq NM_001122955.3) and c.3017A>G (p.(Glu1006Gly)) in KIF1B (RefSeq NM_183416.3) are unlikely pathogenic because they occur in the NHLBI Exome Variant Server (EVS, evs.gs.washington.edu/EVS) with frequencies 0.003 and 0.004, respectively (Figure 2). It can be assumed that dominant mutations causing CMT2 are not frequent in the population because the overall contribution of individual disease mutations in each CMT2 gene is very low. One of the variants, MFN2 c.2113G>A (p.(Val705Ile)) (RefSeq NM_001127660.1), has previously been reported as a CMT2 mutation,10, 11 but we have found that nearly 5% of Finnish individuals are heterozygous carriers of this variant.5 A recent Australian study reported a similar carrier rate and showed that the variant did not segregate with the disease phenotype in a CMT2 family.12 In one patient a potential disease-causing mutation was found in the HSPB1 gene, a c.404C>A transversion (RefSeq NM_001540.3) resulting in p.(Ser135Tyr) amino acid change. This variant was not present in 1000Genomes or EVS database and was predicted to be damaging by the SIFT Genome tool (sift.jcvi.org/). HSPB1 mutations have been reported in autosomal dominant families with axonal CMT or distal hereditary motor neuropathy.13 One of the originally described HSPB1 mutations affected the same amino acid. p.(Ser135), but the change was from serine to phenylalanine (p.(Ser135Phe)) instead of tyrosine. Our patient is from a large family with several affected members and a clear dominant inheritance (Figure 3a). The presence of the mutation was confirmed by Sanger sequencing (Figure 3b), and it segregated with the disease in all nine family members whose samples were available for the analysis.
Data filtering workflow. Sequencing data were filtered with the following steps: (1) from the number of heterozygous variants occurring at least in one patient, the common variants, that is, variants with frequency of 0.005 or higher in the 1000Genomes database, were excluded, (2) exclusion of variants in the non-coding regions that were not intronic variants within 10 bp of the nearest exon, (3) exclusion of coding-region variants that did not change an amino acid. The four remaining variants are described in the table below. The SIFT tool predictions and the frequencies of the variants in the NHLBI ESP EVS are shown.
HSPB1 mutation. (a) The pedigree of the family in which the HSPB1 mutation segregated. The individuals whose DNA samples were sequenced are marked with an asterisk (*). The arrow indicates the index patient who was enrolled in targeted NGS. (b) Sanger sequencing of the index patient confirmed the presence of the mutation, DNA from an unaffected family member was used as control.
In this large family, 18 members in at least 5 generations are known to have axonal motor distal polyneuropathy with minimal sensory involvement. Males have been affected with higher frequency than females (5 female patients compared with 13 males). The disease has been chronic and symmetrical with neurological examination showing onset between the ages of 18 and 25 years in males, and about 10 years later in females. An active sportsman has been an exception with first symptoms occurring at the age of 34 years. Toe and feet extensors were affected the earliest and most prominently, leading to numbness, muscle weakness, peroneus paresis, and finally atrophic paresis of the legs. Fingers and the arms became involved about 5 years after the onset. Older patients had claw hands and needed assistive devices for daily life. All patients finally lost tendon reflexes but had no cranial nerve involvement or autonomic disturbances. Only some of them had mild pes cavus.
The electroneuromyography results (for representative patient, III-5, age 56 years) showed decreased upper limb motor amplitudes in the ulnar and median nerves (0.4 and 0.2 mV, respectively), with marginal decrease in NCV (38.0 and 38.5 m/s, respectively). In the legs, the changes were more pronounced with no detectable measurements in tibial and deep peroneal nerves. Sensory nerve action potential amplitudes were at the lower limit of normal in the radial nerve (9.4 μV), slightly decreased in the median nerve (6.5 μV), and strongly decreased in the ulnar nerve (1.2 μV). Upper limb sensory NCVs were normal in the radial and median nerves (53.0 and 48.7 m/s, respectively), but decreased in the ulnar nerve (28.7 m/s) where the action potential was barely detectable. In electromyography of the lower limb muscles, no activity could be recorded in the tibial or peroneal muscles, whereas the medial head of the gastrocnemius showed fibrillation. Vastus lateralis showed functional deficiency and changes consistent with neurogenic damage. Similar changes were recorded also in the muscles of the hand and lower arm, but the biceps brachii gave almost normal recordings. Another affected family member IV-1, who was studied twice at ages 30 and 32 years showed in the first examination strong and chronic motor nerve axonal damage in distal leg muscles with tibial and deep peroneal nerve amplitudes still detectable, although decreased (0.2 and 0.3 mV, respectively), and NCVs only slightly decreased (37.0 and 34.1 m/s, respectively). Two years later, disease progression was detectable with decrease of motor amplitude in arms and sensory nerve action potential amplitude in legs. Taken together, the findings in this family were consistent with axonal motor-sensory neuropathy, and CMT2 starting in lower leg motor nerves and gradually progressing to the upper limbs and the sensory nerves.
The course of the disease was moderately progressive and most patients in the family required wheelchair after 25–30 years of duration. None of the patients remained fully ambulant and the majority of them used at least lower leg support 5–10 years after the onset. Family members without CMT symptoms were not examined for this study. Subjects were considered unaffected if they reported no symptoms and had passed the typical age of onset.
No potential disease-causing variants were found in the 17 known disease genes in the remaining 12 CMT2 patients. This result suggests that genetic heterogeneity in familial CMT2 may be even larger than previously expected.
Variant analysis in candidate disease genes
We proceeded to analyze the 143 additional candidate genes that were included in the sequencing target either based on association with other neuropathies, functional relation to CMT2 disease genes, or because variants of undetermined pathogenicity had been found in a previous exome sequencing study that included patients from two AD-CMT2 families (Supplementary Table 1). For variant filtering, the following exclusion steps were employed: (1) common variants (1000Genomes frequency <0.005), (2) non-coding variants, (3) synonymous variants, and (4) variants with a non-damaging prediction using the SIFT tool. In the genes that were included because of previous association with related neuropathy or because of functional relationship to the CMT2 disease genes, we found a total of 10 rare, amino-acid altering variants with damaging prediction (Supplementary Table 2). These variants were evaluated for their likelihood of being pathogenic. We used the EVS, which contains data from more than 12 000 exomes of American origin, and 1000Genomes, which contains genome sequences of 100 Finnish individuals among others, to identify mutations present at very low frequency in the population. Of the 10 variants, eight were present in the EVS and/or Finnish 1000Genomes samples, making them unlikely to be pathogenic. The remaining two variants that were not found in the EVS or the Finnish 1000Genomes individuals were c.180G>C (p.(Gln60His)) in PHB2 (RefSeq NM_001144831.1, encoding prohibitin 2), and c.94C>T (p.(Arg32Trp)) in TK2 (RefSeq NM_004614.4, encoding thymidine kinase 2). We screened the TK2 variant by Sanger sequencing and found it to be a common polymorphism after all, as it was present in 5 of our 53 healthy control samples. For the PHB2 variant p.(Gln60His), segregation could not be studied in the corresponding family because additional samples were not available. The mutation was predicted damaging by SIFT, possibly damaging by PolyPhen-2 (genetics.bwh.harvard.edu/pph2), and disease-causing by MutationTaster (www.mutationtaster.org). PHB2 regulates mitochondrial morphology through OPA1-processing and may thus have a role in axon maintenance14 but the pathogenicity of the p.(Gln60His) variant remains presently open.
The c.684C>G (p.(Ile228Met)) variant in the gene SCN9A (RefSeq NM_002977.3), which encodes the voltage-gated sodium channel Naν1.7, was identified in one of our CMT2 families (Supplementary Table 2). Although the EVS frequency of 0.0012 suggests that the variant is likely a rare polymorphism, it has recently been proposed as the causative mutation for patients with small fiber neuropathy. One of the described patients showed severe pain in the teeth, jaw, and behind the eyes as initial symptoms and had normal NCVs, muscle strength, and preserved tendon reflexes.15 His affected sister suffered from pain and redness of the hands and feet triggered by warmth.16 A third patient from another family had asymmetric quantitative sensory test results and marginal loss in the density of intraepidermal nerve fibers. Furthermore, the variant has been shown to impair axon integrity in cultured dorsal root ganglion neurons, and to cause a gain-of-function effect in vitro.17 However, our CMT2 patient carrying the p.(Ile228Met) variant had very different clinical symptoms presenting with bilateral peroneal weakness starting gradually at the age of 35. In the family history, his already deceased father and father’s mother had also had distal leg weakness. At age 40 his electroneuromyography study showed loss in motor nerve amplitudes in legs, and in needle-electromyography chronic neurogenic changes focusing in the distal leg muscles, whereas sensory nerve action potential amplitudes and conduction velocities were within normal limits. During disease progression at age 52 years, motor amplitudes were further decreased and distally absent and sensory nerve action potential amplitudes were decreased in legs while still normal in arms. At this point the patient needed two walking sticks to be able to move. The clinical picture and electroneuromyography findings were consistent with motor-sensory axonal neuropathy. Whether this SCN9A variant could be the underlying cause for CMT2 remains presently open.
Finally, we analyzed the 25 genes that were included as candidates because they had contained potential pathogenic variants in a previous CMT2 exome sequencing study. In these genes, we did not identify any variants that we considered likely to be pathogenic. Unlike the 17 known disease genes, which were our main target, the exons of the additional genes were not Sanger sequenced, although they may have contained uncovered base pairs because of restriction site limitations. Specifically, the available restriction sites allowed inclusion of 99.1% of the desired target, meaning that ∼0.9% of the coding-region bases may have been missed. Thus these genes were not fully excluded for mutations.
Discussion
Molecular diagnosis is challenging to reach in patients with axonal CMT because of a large number of infrequent disease genes, some of which have not yet been discovered. Identification of the causative gene defect allows a precise classification of the disease with establishment of a natural course and prognosis18 and genetic counseling. Treatments have also been proposed, based on targeting the specific pathways that the disease gene is part of.
In our Finnish CMT2 patient cohort, a founder mutation in GDAP1 is a common cause of dominant CMT2, which together with private MFN2 mutations explains up to 25% of the disease.5 The rest of our patients lack genetic diagnosis. This study was motivated by the need to utilize a cost- and time-effective method to exclude known CMT2 disease genes in patients. Our previous studies using WES in dominant CMT2 have resulted either in the identification of a known disease mutation, or in a long list of heterozygous variants without certainty of their pathogenicity. The former end result was desirable but could have been reached with far less sequencing using targeted methods. In the latter result, a new disease gene may be the culprit but we could not rule out all known disease genes because of partial incomplete read coverage. Thus, we tested the efficiency of a targeted NGS approach to identify mutations in the known CMT2 disease genes, or to confidently exclude them.
A recent study described the use of WES for mutation screening in CMT2 patients, identifying disease-causing mutations in known disease genes in 32% of the prescreened sporadic cases.6 This result was highly encouraging, but it required on an average 5930 Mb of sequence per sample. In view of our findings, the same result could have been achieved with targeted NGS of CMT2 candidate genes, requiring just 100 Mb of sequence.
Of the 309 exons in the known 17 CMT2 disease genes, 95% were perfectly covered by the targeted method and this required very little sequencing, considering that the target actually contained an additional 143 genes. In the remaining exons, only a few base pairs were missed in most cases, which reduced the likelihood that mutations would be located in these sites. This was confirmed when we did not find any new variants by Sanger sequencing. For diagnostic purposes, the need to Sanger sequence 5% of the target is not desirable and could be prevented by designing a kit for the target genes with optimized restriction enzymes. The benefits of the targeted method used here are thus the speed and small sequencing requirements, and the limitation is the lack of restriction sites for every target, which requires further optimization.
The analysis of the data was very straightforward and would be feasible in a diagnostic setting. In 17 genes of 13 patients, we identified only 67 different variants altogether. Of these most were easily filtered out as disease mutations because of their frequency in the publicly available genome and exome sequencing databases, which are becoming extremely useful for mutation identification. Although CMT has a prevalence of 1:2500, the contribution of individual mutations in each CMT disease gene is significantly smaller, and thus the chance of finding carriers of dominantly inherited disease mutations in the sequencing databases is low. On the contrary, although also rare, carriers of recessively inherited mutations are more frequent among the individuals whose samples are used as controls for these databases. However, we used caution with the frequency-based exclusion because individuals with late-onset diseases are probable among the control samples. The 1000Genomes data are included in our variant calling pipeline and we used it with a frequency of 0.005. However, at a later stage we excluded the remaining variants using the EVS database, which contains significantly more samples than the 1000Genomes database and allowed thus to exclude variants that were found at frequencies below 0.005. In 1000Genomes, such rare variants may not be listed because by chance they may not have been carried by the sequenced individuals. In a diagnostic analysis of this data, the EVS filter with ∼0.002 frequency would be optimal as the first step and would exclude most polymorphic variants.
NGS methods are well suited for the detection of single nucleotide variants and small insertions or deletions. The technique is relatively less tested for copy number variants involving entire genes or larger genomic regions, although a software has recently been developed for this purpose.19 Unlike CMT1, where PMP22 gene duplication accounts for a significant number of cases,2 the known CMT2 disease mutations so far have been point mutations or small insertions or deletions (IPNMD). In this study, we focused on detection of single nucleotide variants and small insertions or deletions as they were considered to be the most likely mutations in CMT2. Nevertheless, the data analysis could be developed to also detect copy number variants, which were not excluded in our study.
The targeted NGS method proved adept at identifying disease mutations, as we found a new HSPB1 mutation in one large family. The missense mutation changed a conserved serine at position 135 into tyrosine. The p.(Ser135Phe) mutant HSPB1 protein was previously shown to impair cell viability and neurofilament assembly when expressed in cultured cells.13 Tyrosine and phenylalanine both contain a large hydrophobic side chain, suggesting that the substitution of serine to either amino acid has the same consequence and is disease-causing. The p.(Ser135Phe) variant was originally identified in Russian and English families with AD-CMT2F or distal hereditary motor neuropathy, a peripheral neuropathy that closely resembles CMT2 except for the absence of sensory symptoms.13 The clinical phenotype of our patients was similar to the previously described, with progressive symmetrical weakness, atrophy of distal limb muscles initially in the legs and particularly in the peroneal compartment. Furthermore, no sensory symptoms were expressed in our patients, and only in the oldest patients were mild sensory impairment of temperature and vibration in the feet and hands documented upon clinical examination.
Although a number of disease genes have been described for CMT2, it has been estimated that 50% of patients carry mutations in yet unknown disease genes.20 Our results showing complete exclusion of 17 known CMT2 disease genes in 12 unrelated Finnish families with autosomal dominant inheritance supports this hypothesis. With our population history of isolation and bottlenecks, we expect to find new disease genes with founder mutations in these families. The additional 143 candidate genes that were included in the HaloPlex target included genes encoding aminoacyl-tRNA synthetases and proteins involved in axonal transport processes, because of their functional relationship to known CMT2 disease genes. These genes were not fully excluded as disease genes because their NGS gaps were not Sanger sequenced. However, as these NGS restriction site gaps mounted to only 0.9% of the desired target, we can conclude that no obvious disease-causing mutations were found in them. The current repertoire of CMT2 disease genes suggests that the new disease genes can encode proteins of nearly any function and thus the unbiased genome-wide sequencing methods are the choice for disease gene discovery.
In summary, we have introduced and evaluated targeted NGS as a tool to screen disease genes in CMT2. Our results suggest that this technique allows efficient mutation detection and exclusion of candidate genes, which aids in the selection of patients for further genetic studies such as WES or whole-genome sequencing.
References
Szigeti K, Lupski JR : Charcot-Marie-Tooth disease. Eur J Hum Genet 2009; 17: 703–710.
Lupski JR, de Oca-Luna RM, Slaugenhaupt S et al: DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell 1991; 66: 219–232.
Shy ME, Patzko A : Axonal Charcot-Marie-Tooth disease. Curr Opin Neurol 2011; 24: 475–483.
Saporta AS, Sottile SL, Miller LJ, Feely SM, Siskind CE, Shy ME : Charcot-Marie-Tooth disease subtypes and genetic testing strategies. Ann Neurol 2011; 69: 22–33.
Auranen M, Ylikallio E, Toppila J, Somer M, Kiuru-Enari S, Tyynismaa H : Dominant GDAP1 founder mutation is a common cause of axonal Charcot-Marie-Tooth disease in Finland. Neurogenetics 2013; 14: 123–132.
Choi BO, Koo SK, Park MH et al: Exome sequencing is an efficient tool for genetic screening of Charcot-Marie-Tooth Disease. Hum Mutat 2012; 33: 1610–1615.
Lupski JR, Reid JG, Gonzaga-Jauregui C et al: Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med 2010; 362: 1181–1191.
Weedon MN, Hastings R, Caswell R et al: Exome sequencing identifies a DYNC1H1 mutation in a large pedigree with dominant axonal Charcot-Marie-Tooth disease. Am J Hum Genet 2011; 89: 308–312.
Sulonen AM, Ellonen P, Almusa H et al: Comparison of solution-based exome capture methods for next generation sequencing. Genome Biol 2011; 12: R94.
Engelfried K, Vorgerd M, Hagedorn M et al: Charcot-Marie-Tooth neuropathy type 2A: novel mutations in the mitofusin 2 gene (MFN2). BMC Med Genet 2006; 7: 53.
Braathen GJ, Sand JC, Lobato A, Hoyer H, Russell MB : MFN2 point mutations occur in 3.4% of Charcot-Marie-Tooth families. An investigation of 232 Norwegian CMT families. BMC Med Genet 2010; 11: 48.
Albulym OM, Zhu D, Reddel S, Kennerson M, Nicholson G : The MFN2 V705I variant is not a disease-causing mutation: a segregation analysis in a CMT2 family. J Neurodegenerative Dis 2013; 2013: doi:10.1155/2013/495873.
Evgrafov OV, Mersiyanova I, Irobi J et al: Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat Genet 2004; 36: 602–606.
Osman C, Merkwirth C, Langer T : Prohibitins and the functional compartmentalization of mitochondrial membranes. J Cell Sci 2009; 122: 3823–3830.
Faber CG, Hoeijmakers JG, Ahn HS et al: Gain of function Nanu1.7 mutations in idiopathic small fiber neuropathy. Ann Neurol 2012; 71: 26–39.
Estacion M, Han C, Choi JS et al: Intra- and interfamily phenotypic diversity in pain syndromes associated with a gain-of-function variant of NaV1.7. Mol Pain 2011; 7: 92.
Persson AK, Liu S, Faber CG, Merkies IS, Black JA, Waxman SG : Neuropathy-associated Nav1.7 variant I228M impairs integrity of dorsal root ganglion neuron axons. Ann Neurol 2013; 73: 140–145.
Patzko A, Shy ME : Update on Charcot-Marie-Tooth disease. Curr Neurol Neurosci Rep 2011; 11: 78–88.
Sathirapongsasuti JF, Lee H, Horst BA et al: Exome sequencing-based copy-number variation and loss of heterozygosity detection: ExomeCNV. Bioinformatics 2011; 27: 2648–2654.
Braathen GJ, Sand JC, Lobato A, Hoyer H, Russell MB : Genetic epidemiology of Charcot-Marie-Tooth in the general population. Eur J Neurol 2011; 18: 39–48.
Acknowledgements
We thank the patients and their families for their participation in the study. Riitta Lehtinen is thanked for technical help. We also acknowledge the target enrichment, sequencing, and variant calling pipeline analysis performed by the Institute for Molecular Medicine Finland (FIMM). We thank the following funding sources for support: Sigrid Jusélius Foundation, the Finnish Neuromuscular Disorders Association, University of Helsinki and the Academy of Finland, Arvid and Greta Olin’s Foundation, and Finska Läkaresällskapet.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Ylikallio, E., Johari, M., Konovalova, S. et al. Targeted next-generation sequencing reveals further genetic heterogeneity in axonal Charcot–Marie–Tooth neuropathy and a mutation in HSPB1. Eur J Hum Genet 22, 522–527 (2014). https://doi.org/10.1038/ejhg.2013.190
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2013.190
Keywords
This article is cited by
-
Small heat shock proteins in neurodegenerative diseases
Cell Stress and Chaperones (2020)
-
A PRPH splice-donor variant associates with reduced sural nerve amplitude and risk of peripheral neuropathy
Nature Communications (2019)
-
The Variant p.(Arg183Trp) in SPTLC2 Causes Late-Onset Hereditary Sensory Neuropathy
NeuroMolecular Medicine (2016)
-
Dominant transmission of de novo KIF1A motor domain variant underlying pure spastic paraplegia
European Journal of Human Genetics (2015)
-
Ser135Phe mutation in HSPB1 (HSP27) from Charcot–Marie–Tooth disease type 2F families
Genes & Genomics (2015)