Abstract
Nine affected individuals with isolated anophthalmia/microphthalmia from a large Muslim-inbred kindred were investigated. Assuming autosomal-recessive mode of inheritance, whole-genome linkage analysis, on DNA samples from four affected individuals, was undertaken. Homozygosity mapping techniques were employed and a 1.5-Mbp region, homozygous in all affected individuals, was delineated. The region contained nine genes, one of which, aldehyde dehydrogenase 1 (ALDH1A3), was a clear candidate. This gene seems to encode a key enzyme in the formation of a retinoic-acid gradient along the dorsoventral axis during an early eye development and the development of the olfactory system. Sanger sequence analysis revealed a missense mutation, causing a substitution of valine (Val) to methionine (Met) at position 71. Analyzing the p.Val71Met missense mutation using standard open access software (MutationTaster online, PolyPhen, SIFT/PROVEAN) predicts this variant to be damaging. Enzymatic activity, studied in vitro, showed no changes between the mutated and the wild-type ALDH1A3 protein.
Similar content being viewed by others
Introduction
Anophthalmia and microphthalmia (A/M) are rare developmental anomalies resulting in absent or small ocular globes, respectively. Anophthalmia refers to complete absence of the ocular globe in the presence of adnexa (eyelids, conjunctiva and lacrimal apparatus). Microphthalmia is defined as such when the total axial length of the globe is at least two SD below the mean for age; this typically correlates to an axial length below 21 mm in adult eyes.1, 2
A/M is a heterogenous condition. A/M may affect one or both eyes and present as either an isolated or syndromic disorder.1, 2 Both genetic and environmental factors, such as fetal alcohol syndrome and vitamin A excess or deficiency, contribute to congenital eye defects.3, 4, 5, 6, 7 All types of Mendelian inheritance have been reported for A/M. The majority of genes known to be involved in the pathogenesis of A/M are transcription factors or homeobox genes.8
We have previously described a highly inbred Muslim kindred, from Northern Israel, with isolated A/M.9 Nine affected individuals had isolated bilateral microphthalmia and no light perception. All showed normal intelligence, displayed no other anomalies and were otherwise healthy. Responses on electroretinogram or visually evoked potentials were consistently absent in all cases. Computed tomography scans disclosed small congenital cystic globes, with rudimentary optic nerves and extraocular muscles. This study aimed to unravel the genetic basis underlying A/M in our family.
Methods
Patients and families
Four highly inbred nuclear families of Muslim origin displaying autosomal-recessive, isolated A/M were studied. Thirteen healthy parents and siblings and nine affected individuals were clinically investigated. Blood samples were drawn from all participants. Skin biopsy was obtained from one affected individual. The study was approved by the institutional review board at Rambam Health Care Campus, Haifa. Signed informed consent (self and parental) was obtained.
Molecular analyses
Using linkage analyses, known A/M causative genes such as VSX2 (MIM:610093), RAX (MIM:611038), SOX2 (MIM:206900) and PAX6 (OMIM: 607108) were excluded. Genomic DNA of four affected individuals (VI_2, VI_6, VI_12 and VI_13) was genotyped using the Affimetrix GeneChip Human Mapping 250 K Nsp microarray (Santa Clara, CA, USA). Homozygosity-by-descent analysis was carried out manually, exploring identical homozygous intervals in all affected individuals. Linkage analysis, of a candidate homozygous locus on chromosome 15, was expanded to include additional family members, using two microsatellite markers designed by us: GT22 at chr15: 101094111-101094103 (marker_1:F_5′-TGT CAA CCA CGA GCA GTT TC-3′; R_5′-GCC CTC AGC ATC CTG ATA TT-3′) and TG24 at chr15:102147332-102147378 (marker_2:F_5′-GTG GTG GGC TGA TAA ATG CT; R_5′-AAC AAA GGC ATT CTG TGA GGA-3′). Haplotypes of family members were manually constructed and analyzed with SUPERLINK online under the assumption of autosomal-recessive inheritance and disease allele frequency of 0.01. One candidate gene, aldehyde dehydrogenase 1 (ALDH1A3), within the homozygous region was sequenced using genomic DNA from one affected individual. Sanger sequencing of coding regions and flanking intron–exon boundaries was undertaken (primer sequences are available on request). Once the causative mutation was detected (c.211 G>A), a PCR-RFLP assay was developed using primers designed by us: exon3_F:5′-CAG TCT CTC TCT GTT GTT CTG G-3′ and exon3_R:5′-GAG AGC CGT GTC TCA GAG GA-3′. The G to A substitution abolishes an HpyCH4IV restriction-native site, generating cleavage-differential products for the wild-type (277 bp, 35 bp) and the mutated (312 bp) alleles.
Total RNA was extracted from cultured epidermal fibroblasts using the RNeasy Mini Kit (Qiagen, Hilden, Germany) and reverse transcribed into cDNA by the QuantiTect reverse transcription kit (Qiagen) using RT primer mix according to the manufacturer’s protocol. cDNA of ALDH1A3 was sequenced using primers devised by us: cDNA_exon2_F: 5′-GCA ACC TGG AGG TCA AGT TC-3′ and cDNA_exon5_R: 5′-CAG CTT CCA CAC CAG CAT C-3′.
Kinetic studies of the p.Val71Met mutation
ALDH1A3 cDNA was subcloned into the pET-30 Xa/LIC (Novagen, Bloemfontein, South Africa) expression vector, coding for an N-terminal (His)6 tag, by using two primers containing ligation-independent cloning overhangs, as indicated by the manufacturer (EMD Biosciences, Darmstadt, Germany). All reactions were performed in a DNA thermal cycler with Phusion High-Fidelity DNA Polymerase (Thermo Scientific, Waltham, MA, USA). Two mutagenic primers, 5′-GGA GAT AAG CCC GAC ATG GAC AAG GCT GTG GAG GCT GC-3′ and 5′-CTC CAC AGC CTT GTC CAT GTC GGG CTT ATC TCC TTC TTC-3′, were designed to introduce the p.71Met mutation, using the wild-type ALDH1A3 cDNA cloned into pET-30 Xa/LIC as a template. Mutated nucleotides are underlined. The procedure was based on the QuikChange Site-Directed Mutagenesis Kit method (Stratagene, Leicester, UK). The mutagenic strands were sequence verified and then transformed into Escherichia coli BL21 cells, in which recombinant ALDH1A3 was expressed at 24 °C and purified, as previously described10 and as follows: cells were lysed by sonication and the homogenate was applied onto a nickel-charged chelating Sepharose Fast Flow (GE Healthcare, Little Chalfont, UK) column (5 ml). After washing with 60 mM imidazole in 20 mM Tris-HCl, 0.5 M NaCl, pH 8.0, the enzyme was eluted with 250 mM imidazole in the same buffer. Protein fractions were concentrated and imidazole was removed with an Amicon ultra device (Millipore, Little Chalfont, UK). Protein concentration was determined by a dye-binding assay using bovine serum albumin as a standard.11 Enzymatic activity of ALDH1A3 was determined in 50 mM HEPES, 50 mM magnesium chloride, 5 mM dithiothreitol, pH 8.5, at 37 °C, and in the presence of a saturating concentration of cofactor NAD+. The activity with 4-nitrobenzaldehyde was measured in a Cary 400 Bio (Varian, Palo Alto, CA, USA) spectrophotometer, as described.12 Activity with retinoids was analyzed by HPLC; after extraction, retinoids were separated by chromatography on a Nova-Pak Silica column in n-hexane:methyl-tert-butyl ether:isopropanol:acetic acid (95.9:4:0.075:0.025, v/v) mobile phase, at a flow rate of 2 ml/min, and separation of retinaldehyde, retinoic acid and retinol was achieved in a single step, with retention times of 6.76, 8.72 and 29.45 min, respectively, based on previously described protocols.12, 13 An Mr of 244 000 was used to calculate the kcat values. The kinetic constants were expressed as the mean±SEM of two determinations. In addition to HPLC-based kinetics with retinoids, UV–vis spectrophotometric determination of 4-nitrobenzaldehyde oxidation, in which NAD+ reduction was measured at 340 nm, was undertaken.
Results
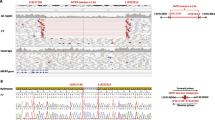
Whole-genome homozygosity mapping defined a single homozygous segment on chromosome 15q26.3 spanning 1 340 206 bp between rs1552101 and rs6598484 shared by four affected individuals. We thereafter developed and genotyped two microsatellite markers in this interval; all affected individuals (n=9) shared the same homozygote haplotype for marker_1, all parents (n=8) were heterozygote for the linked haplotype and healthy siblings (n=5) were either heterozygote or noncarriers of the linked haplotype (Figure 1a). A two-point LOD score of 7.9 was calculated. The region encompassed nine genes, one of which, ALDH1A3 (NM_000693.2 (MIM600463)), a key enzyme in the formation of a retinoic-acid gradient along the dorsoventral axis during an early eye development, was sequenced. Sanger sequencing revealed a homozygous missense mutation c.211G>A (p.Val71Met) in an affected individual (Figure 1b). Using PCR-RFLP analyses, the mutation was tested in all parents and healthy siblings from our four related families and was found to cosegregate with the disease; all affected individuals were homozygous for the mutation (n=9; probability for a random amino-acid exchange 3.3147E-06). Unaffected siblings were either heterozygous or noncarriers (n=5), and all parents were heterozygotes (n=8). The mutation was tested and not observed in 100 individuals from the same ethnic origin.
Haplotypes and mutation c.211G>A in exon 3 of the ALDH1A3 gene. (a) Disease-associated haplotypes are shown in boxes. Markers 1 and rs2573582 define the minimal homozygosity locus associated with the disease (allele 0: not genotyped). (b) The ALDH1A3 c.211G>A (p.Val71Met) mutation in genomic DNA of A/M homozygote individual and carrier.
The mutation was not recorded in the dbSNP databases or the Exome Variant Server. MutationTaster online predicts p.Val71Met to be disease causing (probability score: 0.92). SIFT/PROVEAN (San Diego, CA, USA) and PolyPhen2 predict the mutation to be damaging. ALDH1A3 consists of 13 exons that encode retinaldehyde dehydrogenase, a protein of 512 amino-acid residues involved in retinoic-acid synthesis. Residue 71 is positioned in exon 3, within a DXXDXD motif, between two strictly conserved Asp residues.
The mutation is located at position +7 of exon 3 and does not overlap with the consensus splicing sequence. We, however, have excluded an eventual splicing effect by sequencing the corresponding cDNA from an affected individual. No aberration was identified other than the homozygote G>A substitution.
Wild-type and p.71Met-mutant ALDH1A3 were expressed as soluble proteins in E. coli. Comparable expression levels were observed for the wild-type and p.71Met-mutant ALDH1A3, as assessed from the SDS-PAGE analysis of E. coli lysates (Figure 2). Similar purification yields and activity loss over time were obtained for the mutant and wild-type protein. Under the purification and storage conditions used, both proteins displayed low stability, and precipitation over time was equivalent. No evidence of misfolding or increased instability was observed for the p.71Met mutant.
SDS-PAGE analysis. Fractions obtained during the purification of wild type (lanes 1–3) and p.71M ALDH1A3 (lanes 4–6). Protein was stained with Coomassie Brilliant Blue (Bio Rad, Hercules, CA, USA). Lanes 1 and 4, E. coli lysate (20 μg); lanes 2 and 5, flowthrough from nickel-charged chelating Sepharose; lanes 3 and 6, fractions eluted with 250 mM imidazole.
Kinetic constants were determined using freshly prepared protein. Both wild-type and p.71Met mutant showed comparable enzymatic activity using NAD+ as a cofactor. Kinetic analysis revealed that wild-type and mutant protein display similar Km (0.083±0.01 vs 0.085±0.01 μ M) and kcat (0.79±0.02 vs 0.62±0.02 per min) values for all-trans-retinaldehyde. Very small differences in the Km values for NAD+ (7.9±1.7 vs 4±0.3 μ M) were observed between the wild-type and the mutant enzymes.
UV–vis spectrophotometric determination yielded Vmax values for wild-type and p.71Met-mutant ALDH1A3 of 0.23 and 0.13 μmols per min per mg, respectively.
Discussion
Nine individuals, from one inbred kindred, segregating isolated A/M with orbital cysts were homozygous for a missense mutation in the gene encoding the A3 isoform of the ALDH1A3. The probability for this being a random association stands at 3.8 × 10−6. ALDH1A3 is a key enzyme in the formation of a retinoic-acid gradient along the dorsoventral axis during an early eye development.3, 6, 7 Fares-Taie et al14 have recently shown that mutations in ALDH1A3 cause A/M. Two missense mutations and one splice mutation, namely, p.Arg89Cys, p.Ala493Pro and c.475+1G->T, were reported to segregate in three consanguineous families with A/M. Here, we report on yet another ALDH1A3 missense mutation, namely, p.Val71Met that causes A/M. Although the role of retinoic-acid signaling in the eye development is well established, the results provided by Fares-Taie et al14 and that of ours ascertain the genetic linkage between retinoic-acid synthesis dysfunction and an early eye development anomalies, namely, A/M, in humans.
The mutation described here affects an amino-acid residue that is fairly well conserved in the ALDH1A1 gene orthologs, as well as in the ALDH1A2 and ALDH1A3 gene paralogs. Otherwise, valine (Val) is replaced by another apolar residue (Ile) in most instances. Thr, an isosteric residue, has also been observed to substitute Val (Figure 3). After assessing the highly similar 3D structures of sheep ALDH1A1 (PDB code 1BXS) and rat ALDH1A2 (PDB code 1BI9), we conclude that residue 71 is located at the beginning of an α-helix in the NAD+-binding domain near the protein surface. Apparently, the p.Val71Met substitution should not greatly disturb protein stability or enzymatic function. Methionine (Met) is a rather rare amino acid in proteins, moderately hydrophobic, that similar to Val tends to stabilize α-helices. Unlike residues 89 and 493, reported by Fares-Taie et al,14 Val71 is not involved in subunit–subunit interactions.
Our kinetic assays delivered comparable expression and activity levels for the wild-type and mutant ALDH1A3 proteins. To our knowledge, this report is the first to provide kinetic constants of human ALDH1A3 and retinaldehyde. It has been reported that oxidative stress may render Met susceptible to oxidation, in the process yielding hydrophilic Met sulfoxide and Met sulfone, which can destabilize α-helices. Solvent-exposed Met residues are much more susceptible to oxidation than buried ones, and this may lead to protein misfolding, lower thermal stability or altered activity.15 It is conceivable that the reductive environment of the E. coli cytosol and the low growth temperature in vitro prevented the oxidation and subsequent instability of the recombinant enzyme. It is also likely that conditions used during the purification and enzymatic assay protected Met from being oxidized. Thus, enhanced sensitivity of the p.71Met mutant to oxidation, if the case, might have passed undetected in our assays. Val to Met substitutions were indeed reported to increase temperature sensitivity in other proteins.16, 17 Likewise, a p.Thr49Met mutation in retinol dehydrogenase 12, which reduces retinaldehyde to retinol, was reported to cause severe retinal degeneration.18 This protein, which retained significant catalytic activity in vitro, demonstrated higher instability linked to proteosome degradation in the cell.18
The dynamics of the p.Val71Met mutation remain elusive at this stage. It is plausible that the Met to Val substitution at codon 71 may constitute a secondary initiation for translation mechanisms, or alternately the mutation may cause abnormal protein folding. Further studies on thermal stability, protease sensitivity or in vivo turnover rate of the p.71Met mutant could provide additional information.
Microphthalmia, a strong manifestation within the fetal alcohol spectrum disorder,19 is attributed to a competitive effect of ethanol, or its metabolite acetaldehyde, for retinaldehyde dehydrogenase.20, 21 We could assume that A/M, caused by defects in ALDH1A3, is the genetic alternative of this disorder.
Thus far, the mutations described have been associated with nonsyndromic A/M. The families reported by Fares-Taie et al14 have inconsistently demonstrated additional features (autism and heart anomalies), which may be mutation related or incidental. Fares-Taie et al14 question the validity of these additional signs. In our families, all affected cases had isolated A/M and normal intelligence. Recently, homozygosity for two nonsense mutations in ALDH1A3, namely, c.568A>G (p.Lys190*) and c.1165A>T (p.Lys389*), that predict nonsense-mediated decay and complete loss of function were linked to anophthalmia and hypoplasia of the optic nerve and optic chiasma. With this in mind it remains speculative whether missense mutations are preferentially associated with microphthalmia, whereas nonsense and frameshift mutations are linked to anophtalmia.22
References
Verma AS, Fitzpatrick DR : Anophthalmia and microphthalmia. Orphanet J Rare Dis 2007; 2: 47.
Schittkowski MP, Guthoff RF : Systemic and ophthalmological anomalies in congenital anophthalmic or microphthalmic patients. Br J Ophthalmol 2010; 94: 487–493.
Lee LM, Leung CY, Tang WW et al: A paradoxical teratogenic mechanism for retinoic acid. Proc Natl Acad Sci USA 2012; 109: 13668–13673.
Hornby SJ, Ward SJ, Gilbert CE, Dandona L, Foster A, Jones RB : Environmental risk factors in congenital malformations of the eye. Ann Trop Paediatr 2002; 22: 67–77.
Yelin R, Schyr RB, Kot H et al: Ethanol exposure affects gene expression in the embryonic organizer and reduces retinoic acid levels. Dev Biol 2005; 279: 193–204.
Duester G : Keeping an eye on retinoic acid signaling during eye development. Chem Biol Interact 2009; 178: 178–181.
Molotkov A, Molotkova N, Duester G : Retinoic acid guides eye morphogenetic movements via paracrine signaling but is unnecessary for retinal dorsoventral patterning. Development 2006; 133: 1901–1910.
Bardakjian TM, Schneider A : The genetics of anophthalmia and microphthalmia. Curr Opin Ophthalmol 2011; 22: 309–313.
Porges Y, Gershoni-Baruch R, Leibu R et al: Hereditary microphthalmia with colobomatous cyst. Am J Ophthalmol 1992; 114: 3.
Ruiz FX, Porté S, Gallego O et al: Retinaldehyde is a substrate for human aldo-keto reductases of the 1C subfamily. Biochem J 2011; 440: 335–344.
Bradford MM : A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976; 72: 248–254.
Gallego O, Belyaeva OV, Porté S et al: Kinetic Analysis of SDRs, ADHs and AKRs toward Free and CRBPI-bound Retinoids: Effect of Tween-80 and Microsomal Membranes; in Weiner H, Lindahl R, Maser E, Plapp B (eds): Enzymology and Molecular Biology of Carbonyl Metabolism 13. West Lafayette, IN, USA: Purdue University Press, 2007, pp 144–151.
Barua AB : Improved normal-phase and reversed-phase gradient high-performance liquid chromatography procedures for the analysis of retinoids and carotenoids in human serum, plant and animal tissues. J Chromatogr A 2001; 936: 83.
Fares-Taie L, Gerber S, Chassaing N et al: ALDH1A3 mutations cause recessive anophthalmia and microphthalmia. Am J Hum Genet 2013; 92: 265–270.
Wolschner C, Giese A, Kretzschmar HA, Huber R, Moroder L, Budisa N : Design of anti- and pro-aggregation variants to assess the effects of methionine oxidation in human prion protein. Proc Natl Acad Sci USA 2009; 106: 7756–7761.
Egan MF, Goldberg TE, Kolachana BS et al: Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci USA 2001; 98: 6917–6922.
Egan MF, Kojima M, Callicott JH et al: The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell 2003; 112: 257–269.
Lee SA, Belyaeva OV, Kedishvili NY : Disease-associated variants of microsomal retinol dehydrogenase 12 (RDH12) are degraded at mutant-specific rates. FEBS Lett 2010; 584: 507–510.
Strömland K, Pinazo-Durán MD : Ophthalmic involvement in the fetal alcohol syndrome: clinical and animal model studies. Alcohol Alcohol 2002; 37: 2–8.
Kot-Leibovich H, Fainsod A : Ethanol induces embryonic malformations by competing for retinaldehyde dehydrogenase activity during vertebrate gastrulation. Dis Model Mech 2009; 2: 295–305.
Dupé V, Matt N, Garnier JM, Chambon P, Mark M, Ghyselinck NB : A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proc Natl Acad Sci USA 2003; 100: 14036–14041.
Yahyavi M, Abouzeid H, Gawdat G et al: ALDH1A3 loss of function causes bilateral anophthalmia/microphthalmia and hypoplasia of the optic nerve and optic chiasm. Hum Mol Genet 2013; 22: 3250–3258.
Acknowledgements
We are grateful to all family members for their participation in this study. We thank Gregg Duester who provided the ALDH1A3 cDNA clone. Affymetrix GeneChip Human Mapping 250K Nsp microarray was carried out in the Biological Services Unit at the Weizmann Institute. This work was supported in part by the Spanish Ministerio de Economía y Competitividad (BFU2011-24176) and Generalitat de Catalunya (2009 SGR 795).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Mory, A., Ruiz, F., Dagan, E. et al. A missense mutation in ALDH1A3 causes isolated microphthalmia/anophthalmia in nine individuals from an inbred Muslim kindred. Eur J Hum Genet 22, 419–422 (2014). https://doi.org/10.1038/ejhg.2013.157
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2013.157
Keywords
This article is cited by
-
Genetics and functions of the retinoic acid pathway, with special emphasis on the eye
Human Genomics (2019)
-
Genetic architecture of retinoic-acid signaling-associated ocular developmental defects
Human Genetics (2019)
-
Genetics of anophthalmia and microphthalmia. Part 1: Non-syndromic anophthalmia/microphthalmia
Human Genetics (2019)
-
Novel mutations in ALDH1A3 associated with autosomal recessive anophthalmia/microphthalmia, and review of the literature
BMC Medical Genetics (2018)
-
Genetic Testing for Eye Diseases: A Comprehensive Guide and Review of Ocular Genetic Manifestations from Anterior Segment Malformation to Retinal Dystrophy
Current Genetic Medicine Reports (2016)