Abstract
Although many genes have been identified for the autosomal recessive cerebellar ataxias (ARCAs), several patients are unlinked to the respective loci, suggesting further genetic heterogeneity. We combined homozygosity mapping and exome sequencing in a consanguineous Egyptian family with congenital ARCA, mental retardation and pyramidal signs. A homozygous 5-bp deletion in SPTBN2, the gene whose in-frame mutations cause autosomal dominant spinocerebellar ataxia type 5, was shown to segregate with ataxia in the family. Our findings are compatible with the concept of truncating SPTBN2 mutations acting recessively, which is supported by disease expression in homozygous, but not heterozygous, knockout mice, ataxia in Beagle dogs with a homozygous frameshift mutation and, very recently, a homozygous SPTBN2 nonsense mutation underlying infantile ataxia and psychomotor delay in a human family. As there was no evidence for mutations in 23 additional consanguineous families, SPTBN2-related ARCA is probably rare.
Similar content being viewed by others
Introduction
Genetic ataxias are inherited as autosomal dominant, recessive, X-linked or mitochondrial traits. Autosomal recessive cerebellar ataxias (ARCAs; incidence: ∼5/100 000) have to be considered in patients younger than 30 years. Infantile onset is common, and patients with congenital variants typically have developmental delay. At least 20 genes have been reported, but about 40% of cases are due to mutations in yet unidentified genes.1, 2
Materials and methods
See Supplementary Information. The SPTBN2 mutation described in this manuscript has been submitted to the Leiden Open Variation Database (LOVD v.3.0. Build 05).
Results
Clinical presentation
II:1 is a 9-year-old girl, the first child of healthy consanguineous parents. She was born full term after uncomplicated pregnancy by normal vaginal delivery. At the age of 9 years, she was referred to the genetic clinic with global developmental delay and ataxia. At 9 months, her mother noticed that she could not sit unsupported. Her motor skills developed but with a significant delay. She could walk at the age of 2.5 years, but her gait was wide-based, and tremors occurred when eating or holding toys. She received multivitamins, vitamin D and physiotherapy. According to the parents, her condition slowly improved: She is able to walk and eat without support now, although with difficulties. On examination, her height, weight and skull circumference were normal. She had global developmental delay and no dysmorphic features. Speech was slurred. Chest, heart and abdominal examination gave normal results. Neurological examination revealed no cranial nerve affection, nystagmus or muscle atrophy. Muscular strength could not be examined because the patient did not cooperate, but muscular tone appeared increased (spasticity with clasp-knife response). Deep tendon reflexes were increased with extensor plantar response (positive Babinski sign). Superficial reflexes (ipsilateral contraction of upper and lower abdominal muscles upon stroke) were lost, whereas sensation was normal. Signs of cerebellar dysfunction included dysmetria, intention tremors and dysdiadochokinesia. There were no extrapyramidal manifestations or bulbar symptoms. No cranial CT or MRI was available. The younger brother of II:1, II:2, was similarly affected with global developmental delay, clasp-knife spasticity (with increased deep tendon reflexes, positive Babinski sign) and cerebellar dysfunction (with intention tremors, dysmetria and ataxic gait). The family history revealed a similarly affected maternal cousin born to healthy consanguineous parents, too (Figure 1a).
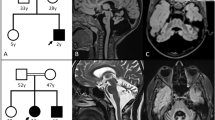
(a) Pedigree of the ARCA family investigated herein. DNA from patients with numbers were available for genetic analysis. The sample of patient II:2 was submitted to exome sequencing. (b) Graphical view of the LOD score calculation of genome-wide SNP mapping. Regions showing HBD were identified on chromosomes 2 (2 times), 5, 6, 9, 10 and 11 and are indicated by arrows (red arrow: disease gene region comprising SPTBN2 on chromosome 11). (c) Sanger sequencing confirmed the homozygous mutation, c.2864_2868del (p.(Thr955Serfs*120)) in SPTBN2 exon 16, in both patients (middle panel). It was found in heterozygous state in both parents (lower panel). Upper panel: wild-type sequence from a healthy control. (d) Scheme of SPTBN2 gene structure and deduced protein with locations of the recessive (in red; human mutations described herein and by Lise et al.11, and the more C-terminal mutation that was recently identified in Beagle dogs) and dominant (SCA5; in blue) mutations. For clarity, the one-letter code was applied for mutation designation in this subset of the figure.
Genetic analysis
We identified seven regions of homozygosity by descent (HBD) with a combined maximum parametric LOD score of 2.783 on chromosomes 2, 5, 6, 9, 10 and 11 (Figure 1b; Supplementary Figure S1). Exome sequencing revealed 424 genes with novel, homozygous variants that resulted in stop codons gained, stop codons lost, frameshifts, non-synonymous coding changes and essential splice site mutations. Sixty-seven of these are contained in the chromosome regions with maximum significance, 43 of which are contained in the largest HBD region on chromosome 11 (∼50 Mb). Statistically, the homozygous disease-causing mutation in the offspring of consanguineous parents most likely resides in the region defined by the longest run of homozygous SNPs.3 In this region, 3 genes carried essential splice site variants, 7 genes had frameshift coding deletions and 12 genes had non-synonymous variants with aggregated SIFT, Polyphen and Condel predictions for deleterious effects. Excluding anonymous ORF and olfactory receptor genes and prioritizing the frameshift variants over non-synonymous and splice site changes left four genes with homozygous frameshift variants, SPTBN2, B3GNT6, SF1 and RELT. All of these are single-base insertions, likely homopolymer misalignments or at low (<6 times) coverage, with the exception of the mutation in SPTBN2 (16 times coverage). SPTBN2 (MIM number 604985) carries a c.2864_2868del (p.(Thr955Serfs*120)) in exon 16 (Supplementary Figure S2) and was of particular interest because SPTBN2 is mutated in spinocerebellar ataxia type 5 (SCA5; MIM number 600224). B3GNT6 codes for an enzyme involved in the biosynthesis of mucin-type glycoproteins in the digestive tract. SF1 codes for a splicing factor and has, although known since 1984, as yet not been implicated in any human disorder. RELT designates the gene for a protein from the tumor necrosis factor receptor superfamily, is expressed in lymphoid tissue and induces NF-κβ activation. Sanger sequencing of SPTBN2 confirmed the mutation in homozygous state in II:1. The affected brother, II:2, was also homozygous for this mutation, while both parents were heterozygous (Figure 1c). The mutation predicts a truncated protein of 1074 residues (wildtype: 2390 residues) including 119 unrelated residues (Supplementary Figure S3). In two small families out of 23 consanguineous families with sporadic or apparently recessive cerebellar ataxia (including 16 from Egypt), index patients displayed homozygous alleles for all three SPTBN2-flanking markers, but also heterozygosity for a presumably nonpathogenic variant in the 3′-prime UTR (c.*89C>G, both cases) and for a common SNP (rs4930388, one case). Thus, we discontinued SPTBN2 sequencing after 32 and 25 exons, respectively.
Discussion
Spectrins are scaffolding proteins and important structural components of the plasma membrane skeleton, controlling its shape, organization and integrity. They participate in the transport of cellular organelles and help assembling specialized membrane domains. SPTBN2 encodes a β3-spectrin with high expression in Purkinje cells that is involved in excitatory glutamate signaling through stabilization of the glutamate transporter EAAT4 at the membrane’s surface. Heterozygous in-frame SPTBN2 mutations cause SCA5, a rare SCA subtype with only five mutations reported,4, 5, 6 with the mean age of onset at 33 years and mostly normal lifespan.
We describe a homozygous 5-bp deletion in SPTBN2 in a consanguineous family with infantile ataxia, developmental delay and pyramidal signs that was unlinked to the known ARCA loci. The differences between dominant and recessive SPTBN2-linked ataxia resemble genotype-phenotype correlations in other genetically heterogeneous conditions, for example, deafness: biallelic MYO6 mutations cause profound congenital deafness, DFNB37,7 whereas heterozygous mutations manifest as progressive postlingual hearing loss, DFNA22.8 A single SCA5 patient has been reported with a heterozygous missense SPTBN2 mutation and an infantile onset, resembling the phenotype in our family.6 Her mutation affects a highly conserved protein residue (p.Arg480Trp). Arg480 may represent a residue of particular importance for SPTBN2 function, with an unusually severe phenotype when mutated. However, in view of the clinical picture of SCA5 reported so far and regarding our findings, either a second-site SCA5 modifier or an undetected secondary (recessive) SPTBN2 mutation in trans (eg, deep intronic or in a non-coding regulatory region) contributing to disease appears likely in this early-onset case.
The SPTBN2 mutation in our family is likely causative for several reasons: The early frameshift predicts either an unstable RNA or a likely non-functional protein that is less than half of the wildtype in size (Figure 1d and Supplementary Figure S3). Moreover, loss of one Sptbn2 copy was tolerated in mice, whereas homozygous Sptbn2 knockout mice displayed ataxia,9 suggesting a recessive effect of loss-of-function mutations. Compatible with these findings, exome sequencing in Beagle dogs with autosomal recessive neonatal cerebellar cortical degeneration recently identified a homozygous Sptbn2 frameshift mutation.10 Both homozygous knockout mice and Beagle dogs show Purkinje cell loss and cerebellar atrophy with molecular layer thinning. Sptbn2 deficiency in mice leads to a Purkinje cell membrane defect due to reduced sodium currents and altered glutamate signaling.9 It is noteworthy that a parallel independent study identified a homozygous SPTBN2 nonsense mutation in a human family with infantile ataxia and psychomotor delay. The patients in this study showed clear cerebellar atrophy.11 No cranial MRIs were available for our patients; although cerebellar symptoms do not necessarily correlate with cerebellar atrophy, we assume that they have cerebellar atrophy too.
We largely excluded a causative role of SPTBN2 in 23 additional consanguineous families, indicating that SPTBN2 is probably a rare ARCA gene. Our study confirms that autosomal dominant and recessive SCA may be allelic disorders. As a consequence, the complete coding sequences of genes involved in dominant SCA types, mostly pathogenic through expansion of CAG triplets, should be considered in unresolved cases of recessive ataxia and vice versa.
References
Anheim M, Tranchant C, Koenig M : The autosomal recessive cerebellar ataxias. N Engl J Med 2012; 366: 636–646.
Sailer A, Houlden H : Recent advances in the genetics of cerebellar ataxias. Curr Neurol Neurosci Rep 2012; 12: 227–236.
Woods CG, Cox J, Springell K et al: Quantification of homozygosity in consanguineous individuals with autosomal recessive disease. Am J Hum Genet 2006; 78: 889–896.
Ikeda Y, Dick KA, Weatherspoon MR et al: Spectrin mutations cause spinocerebellar ataxia type 5. Nat Genet 2006; 38: 184–190.
Cho E, Fogel BL : A family with spinocerebellar ataxia type 5 found to have a novel missense mutation within a SPTBN2 spectrin repeat. Cerebellum 2012; 12: 162–164.
Jacob FD, Ho ES, Martinez-Ojeda M, Darras BT, Khwaja OS : Case of infantile onset spinocerebellar ataxia type 5. J Child Neurol 2012, e-pub ahead of print 21 August 2012; doi:10.1177/0883073812454331.
Ahmed ZM, Morell RJ, Riazuddin S et al: Mutations of MYO6 are associated with recessive deafness, DFNB37. Am J Hum Genet 2003; 72: 1315–1322.
Melchionda S, Ahituv N, Bisceglia L et al: MYO6, the human homologue of the gene responsible for deafness in Snell’s waltzer mice, is mutated in autosomal dominant nonsyndromic hearing loss. Am J Hum Genet 2001; 69: 635–640.
Perkins EM, Clarkson YL, Sabatier N et al: Loss of beta-III spectrin leads to Purkinje cell dysfunction recapitulating the behavior and neuropathology of spinocerebellar ataxia type 5 in humans. J Neurosci 2010; 30: 4857–4867.
Forman OP, De Risio L, Stewart J, Mellersh CS, Beltran E : Genome-wide mRNA sequencing of a single canine cerebellar cortical degeneration case leads to the identification of a disease associated SPTBN2 mutation. BMC Genet 2012; 13: 55.
Lise S, Clarkson Y, Perkins E et al: Recessive mutations in SPTBN2 implicate beta-III spectrin in both cognitive and motor development. PLoS Genet 2012; 8: e1003074.
Acknowledgements
We are indebted to the families that participated in this study. HJB was supported by the Deutsche Heredo-Ataxie-Gesellschaft e.V., http://www.ataxie.de. The research leading to these results received funding from the European Community's Seventh Framework Program FP7/2007-2013 under grant agreement 2012-305121 (project acronym NeurOmics) to RH.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
HJB is employee of Bioscientia, which is part of a publicly traded diagnostic company. The remaining authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Elsayed, S., Heller, R., Thoenes, M. et al. Autosomal dominant SCA5 and autosomal recessive infantile SCA are allelic conditions resulting from SPTBN2 mutations. Eur J Hum Genet 22, 286–288 (2014). https://doi.org/10.1038/ejhg.2013.150
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2013.150
Keywords
This article is cited by
-
The inherited cerebellar ataxias: an update
Journal of Neurology (2023)
-
A heterozygous GRID2 mutation in autosomal dominant cerebellar ataxia
Human Genome Variation (2022)
-
Spinocerebellar Ataxia Type 5 (SCA5) Mimicking Cerebral Palsy: a Very Early Onset Autosomal Dominant Hereditary Ataxia
The Cerebellum (2022)
-
Between SCA5 and SCAR14: delineation of the SPTBN2 p.R480W-associated phenotype
European Journal of Human Genetics (2018)
-
A Novel Homozygous Mutation in SPTBN2 Leads to Spinocerebellar Ataxia in a Consanguineous Family: Report of a New Infantile-Onset Case and Brief Review of the Literature
The Cerebellum (2018)