Abstract
Fanconi anemia is a genetically heterogeneous autosomal recessive disorder characterized by development abnormalities, bone marrow failure, and childhood cancers. Compelling evidence indicates a common genetic basis for FA and breast/ovarian cancer susceptibility. Recently, biallelic germ-line mutations in SLX4 have been demonstrated to cause a previously unknown FA subtype (FA-P). We address the role of SLX4/FANCP in breast/ovarian cancer susceptibility by conducting a comprehensive mutation scanning in 486 index cases from non-BRCA1/BRCA2 multiple-case breast and/or ovarian cancer families (non-BRCA1/2 families) from Spain. We detected one unequivocal loss-of-function mutation (p.Glu1517X). In addition, one missense change (p.Arg372Trp) predicted to be pathogenic by in silico analysis co-segregates with disease in one family. Overall, the study indicates that SLX4 mutation screening will have a very low impact (if any) in the genetic counseling of non-BRCA1/2 families.
Similar content being viewed by others
Introduction
BRCA1 and BRCA2 germ-line mutations explain roughly 20% of the multiple-case breast and/or ovarian cancer families attending familial cancer clinics. Non-BRCA1/2 families have been the focus of several genetic studies aimed to identify additional causative loci, albeit with little success.1
Fanconi Anemia (FA) is a genetically heterogeneous rare autosomal recessive disorder characterized by developmental abnormalities, bone marrow failure, and childhood cancers.2 Somehow unexpectedly, a common genetic basis for breast/ovarian cancer susceptibility and FA emerged when biallelic BRCA2 loss-of-function mutations were identified in FA type D1 patients.3 Later, mutations in FANCJ/BRIP1, FANCN/PALB2, FANCO/RAD51C, and XRCC2 have been found to lead both to FA (when biallelic) and to increased breast cancer risk (when monoallelic).4, 5, 6, 7
Recently, two studies have demonstrated that SLX4 loss-of-function biallelic mutations explain a novel FA subgroup (FANCP).8, 9 Not surprisingly, both reports suggest that SLX4 might be a breast and/or ovarian cancer predisposing gene. So far, this hypothesis has been tested by analyzing SLX4 in 52 German/Byelorussian, 94 Spanish and 526 Italian non-BRCA1/2 families, but obvious loss-of-function germ-line mutations (or any other evidence supporting a causative role) have not been reported.10, 11, 12 To further investigate the role of SLX4 in breast/ovarian cancer susceptibility, we have performed a comprehensive scanning of germ-line mutations in a cohort of 486 index cases from Spanish non-BRCA1/2 families.
Patients and methods
486 unrelated women (465 breast and 21 ovarian cancer patients) who were referred for BRCA1 and BRCA2 genetic testing due to a family history of breast and/or ovarian cancer compatible with a hereditary syndrome (a minimum of two additional breast and/or ovarian cancer cases diagnosed at any age in two generations of the same parental branch). After comprehensive genetic testing (full coding sequence, intron/exon boundaries and genomic rearrangements), all 486 women were considered non-BRCA1/2 patients, that is, genetic testing discarded the presence of BRCA1/2 truncating mutations or any other sequence variant described as a disease-causing mutation in the Breast Informative Core (BIC) database (http://research.nhgri.nih.gov/bic/). The MLPA KIT P045 (MRC-Holland) detected the CHEK2*1100delC allelic variant in one woman (later confirmed by direct sequencing). As this is a low-penetrance allele,13 the carrier was not excluded from further analysis. Families were ascertained through three participating centers: Hospital Clinico San Carlos, Madrid, (N=348), Fundación Publica Galega de Medicina Xenomica, Santiago de Compostela, (N=98), and Hospital Vall d’Hebrón, Barcelona (N=40). The study was approved by the Ethics Committee of each participating center. All patients signed an informed consent to participate in the study. DNA samples were obtained from peripheral blood leukocytes using the standard procedures.
A total of 29 SLX4 amplicons were designed (Supplementary Table 1). PCR amplification and high-resolution melting analysis were carried out in a 7500 Fast Real-Time PCR Instrument (Applied Biosystems, Foster City, CA, USA) with Melt-Doctor HRM Master Mix (Applied Biosystems) and default settings. Samples were sequenced using the Big Dye Terminator v1.1 Cycle Sequencing Reaction kit (Applied Biosystems), according to the manufacturer’s protocol. All sequence variants are named according to the Human Genome Variation Society (HGVS) nomenclature, using Ensemble transcript ID ENST00000294008 as a reference sequence. All variants were investigated in dbSNP135 (http://www.ncbi.nlm.nih.gov/SNP/) and Exome Variant Server (http://evs.gs.washington.edu/EVS/), Exome Sequencing Project (ESP), Seattle, WA (Table 1, and Supplementary Table 2).
Splicing predictions were performed with Human Splicing Finder and MaxEnt splice site matrices (http://www.umd.be/HSF/). Discrepant results were analyzed with NNSPLICE (http://www.fruitfly.org/seq_tools/splice.html), and GeneSplicer (http://www.cbcb.umd.edu/software/genesplicer/). Bioinformatic analyses of missense substitutions were performed using PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), SIFT (http://sift.jcvi.org/), Align-GVGD (http://agvgd.iarc.fr/), and Panther (http://www.pantherdb.org/tools/csnp/). In all cases, default settings were applied. For Align-GVGD analysis, we performed Clustal-W multiple-sequence alignment of Homo, Chimpanzee, Cow, Dog, Guinea, Mouse, Opossum, and Zebrafish SLX4 sequences. Statistical analyses were performed with JavaStat (http://statpages.org/).
Results
We used high-resolution melting analysis to investigate the whole coding region and flanking sequences of the SLX4 gene in 486 index cases from non-BRCA1/2 families. Fifty-seven different sequence variants (11 not previously annotated in dbSNP) were identified, including missense (N=31), silent (N=18), intronic (N=6), 5′UTR (N=1) and nonsense (N=1) changes (Supplementary Table 2).
The c.2854G>A (p.Ala952Thr) and c.2855C>T (p.Ala952Val) substitutions are recorded in dbSNP135 as two individual common variants (rs59939128, and rs78637028, respectively). However, we noted that all c.2855C>T carriers were c.2854G>A carriers as well. The most parsimonious model indicates that the double-nucleotide (doublet) substitution c.2854_55GC>AT (p.Ala952Met) is the actual common variant. We identified one true c.2854G>A (p.Ala952Thr) carrier, indicating that this is a rare variant (at least in our population). Similar observations have been made previously.10, 12
Variants with MAF >0.01, and/or variants in which homozygous carriers have been reported (Supplementary Table 2) were considered likely neutral polymorphisms (not clinically relevant) and discarded for further analysis (N=23). The truncating mutation p.Glu1517X (not annotated in dbSNP135) was the only unequivocal loss-of-function SLX4 mutation detected. The mutated allele codifies for a truncated protein lacking 318 residues at the C-terminal end. The deleted region includes SAP (SAF-A/B, Acinus and PIAS) and SBD (SLX1-binding domain) domains critical for proper SLX4 function.14 Unfortunately, samples from other family members were not available, so it was not possible to perform co-segregation analysis (Figure 1). The mutation was not detected in 480 healthy control samples (Table 1). The remaining variants (N=36) were analyzed in silico to evaluate their potential impact on mRNA splicing and/or protein function. Neither HSF nor MaxEntScan predicted the splicing effects for any variant, with the single exception of c.4115G>A (p.Arg1372Gln). This variant was predicted by HSF to create a new donor site. MaxEntScan, NNSPLICE, and GeneSplicer did not predict any effect, so it was considered unlikely that this variant affects normal splicing.
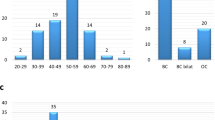
Left panel displays a representative electropherogram demonstrating the SLX4 germ-line mutation c.4549G>T (p.Glu1517X). This truncating mutation was identified in a pedigree (right panel) with multiple cases of breast cancer, but not ovarian cancer. The mutation was identified in the index case (arrow). Unfortunately, it was not possible to perform co-segregation analysis. BC, breast cancer. dx, age at diagnosis.
Eight missense variants were predicted to be functionally relevant by two or more algorithms (Table 1). Because of lack of additional family members, co-segregation analysis was restricted to p.Gly141Trp and p.Arg372Trp families (Figure 2). The p.Gly141Trp variant was detected in one family carrying the CHEK2*1100delC variant as well, thereby complicating co-segregation analysis. The p.Arg372Trp variant co-segregates with disease in two affected family members other than the proband. Four missense variants were tested in a control group and two of these, including p.Arg372Trp, were only found in the patient group (Table 1).
The SLX4 variants p.Arg372Trp and p.Gly141Trp are predicted to be pathogenic by in silico analysis. In the case of p.Arg372Trp, co-segregation analysis is compatible with a pathogenic role (family ID_452). In the case of p.Gly141Trp, co-segregation analysis is complicated by the fact that CHEK2*1100delC is also detected in this family (family ID_1035). Index cases are indicated with arrows. BC, breast cancer. GC, gastric cancer. dx, age at diagnosis.
Discussion
We have detected one unequivocal loss-of-function SLX4 germ-line mutation (p.Glu1517X) in 486 non-BRCA1/2 families tested, confirming that these alterations are rare, but do exist. Previous studies conducted in non-BRCA1/2 families failed to detect such alterations.10, 11, 12 Interestingly, not a single SLX4 truncating (or consensus splice) variant has been observed in >12,000 exomes from the ESP project, supporting the relevance of our finding.
Yet, our analysis provides further evidence supporting that the proportion of non-BRCA1/2 families explained by SLX4 germ-line defects (if any) is probably very low. Indeed, the existence of genes able to explain (individually) a substantial fraction of non-BRCA1/2 families seems unlikely, as genome-wide linkage analysis failed to identify such hypothetical loci.1 Consistently with that, RAD51C and XRCC2 germ-line mutations have been reported in a very low proportion (<1%) of non-BRCA1/2 families.6, 7
Certainly, the actual prevalence of germ-line mutations in those genes might be underestimated due to the fact that non-truncating disease-causing mutations are difficult to distinguish from clinically irrelevant rare variants.15 For instance, we suspected that p.Arg372Trp is a true pathogenic mutation. This is because this variant is predicted to be pathogenic by four algorithms with very high scores, co-segregates with disease, and has not been detected in 480 healthy control individuals (nor in 12,000 exomes from the ESP project, Supplementary Table 2). Moreover, this variant targets the UBZ4 domain (Ubiquiting-Binding Zinc finger motif 4) essential for SLX4 interaction with multiple-partner proteins,14 and recruitment to sites of DNA damage.16 Indeed, four FA patients harbor SLX4 splicing mutations causing in-frame exon 5 skipping,8, 9 supporting that the UBZ4 domain (encoded by exon 5) is clinically relevant. Other variants do not appear to represent disease-causing mutations, in particular those detected in control samples, but some might represent low- or moderate-risk alleles contributing to the overall cancer risk. Interestingly, our cohort included one family harboring both the SLX4 variant p.Gly141Trp, and CHEK2*1100delC. It has been postulated that CHEK2*1100delC might interacts with another rare gene (or genes) to cause breast cancer risk comparable with those conferred by BRCA1 or BRCA2.17 Our data suggest that SLX4 might be one of such rare-interacting genes.
We think that routine analysis of SLX4 in non-BRCA1/2 families by classical technologies such as Sanger sequencing is not warranted. Yet, this and other FA-related genes can be incorporated easily into novel testing strategies who analyze large panels of breast and ovarian cancer susceptibility genes with massively parallel sequencing.18 In this scenario, the identification of people whose breast cancer is related to homologous recombination DNA-repair defects seems clinically relevant, as those patients might benefit from Poly (ADP-ribose) polymerase (PARP) inhibitor therapies.
In conclusion, the prevalence of SLX4 loss-of-function mutations among non-BRCA1/2 families is probably very low. Yet, to definitely demonstrate (or discard) a role for SLX4 in breast and/or ovarian cancer genetic susceptibility is of outstanding scientific interest, so that further studies in very large cohorts are warranted.
References
Oldenburg RA, Meijers-Heijboer H, Cornelisse CJ, Devilee P : Genetic susceptibility for breast cancer: how many more genes to be found? Crit Rev Oncol Hematol 2007; 63: 125–149.
Moldovan GL, D’Andrea AD : How the fanconi anemia pathway guards the genome. Annu Rev Genet 2009; 43: 223–249.
Howlett NG, Taniguchi T, Olson S et al: Biallelic inactivation of BRCA2 in Fanconi anemia. Science 2002; 297: 606–609.
Seal S, Thompson D, Renwick A et al: Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet 2006; 38: 1239–1241.
Rahman N, Seal S, Thompson D et al: PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet 2007; 39: 165–167.
Meindl A, Hellebrand H, Wiek C et al: Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet 2010; 42: 410–414.
Park DJ, Lesueur F, Nguyen-Dumont T et al: Rare mutations in XRCC2 increase the risk of breast cancer. Am J Hum Genet 2012; 90: 734–739.
Stoepker C, Hain K, Schuster B et al: SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet 2011; 43: 138–141.
Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A : Mutations of the SLX4 gene in Fanconi anemia. Nat Genet 2011; 43: 142–146.
Landwehr R, Bogdanova NV, Antonenkova N et al: Mutation analysis of the SLX4/FANCP gene in hereditary breast cancer. Breast Cancer Res Treat 2011; 130: 1021–1028.
Fernández-Rodríguez J, Quiles F, Blanco I et al: Analysis of SLX4/FANCP in non-BRCA1/2-mutated breast cancer families. BMC Cancer 2012; 12: 84.
Catucci I, Colombo M, Verderio P et al: Sequencing analysis of SLX4/FANCP gene in Italian familial breast cancer cases. PLoS One 2012; 7: e31038.
Meijers-Heijboer H, van den Ouweland A, Klijn J et al: CHEK2-Breast Cancer Consortium, low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet 2002; 31: 55–59.
Svendsen JM, Smogorzewska A, Sowa ME et al: Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 2009; 138: 63–77.
Osorio A, Endt D, Fernández F et al: Predominance of pathogenic missense variants in the RAD51C gene occurring in breast and ovarian cancer families. Hum Mol Genet 2012; 21: 2889–2898.
Yamamoto KN, Kobayashi S, Tsuda M et al: Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc Natl Acad Sci USA 2011; 108: 6492–6496.
Oldenburg RA, Kroeze-Jansema K, Kraan J et al: The CHEK2*1100delC variant acts as a breast cancer risk modifier in non-BRCA1/BRCA2 multiple-case families. Cancer Res 2003; 63: 8153–8157.
Walsh T, Lee MK, Casadei S et al: Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc Natl Acad Sci USA 2010; 107: 12629–12633.
Acknowledgements
We thank the patients whose participation made this project possible. We thank Paula Diaque for technical assistance. This study was supported by Instituto de Salud Carlos III, Fondo de Investigación Sanitaria (FIS) Research Grants 09/00859, and Fundación Mutua Madrileña (FMM) Research Grant FMM-08, to MH. Xunta de Galicia (10PXIB 9101297PR) and Fundación Mutua Madrileña (FMM-10) to AV. Red Temática de Investigación Cooperativa en Cáncer; Instituto de Salud Carlos III, Fondo Europeo de Desarrollo Regional (RETICC 06/0020/0021) supported PG, AR, PPS, EDR, TC, and MdH. None of these study sponsors had any role in study design, in the collection, analysis, and interpretation of data; in the writing of the report; and/or in the decision to submit the paper for publication.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
de Garibay, G., Díaz, A., Gaviña, B. et al. Low prevalence of SLX4 loss-of-function mutations in non-BRCA1/2 breast and/or ovarian cancer families. Eur J Hum Genet 21, 883–886 (2013). https://doi.org/10.1038/ejhg.2012.268
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2012.268
Keywords
This article is cited by
-
Expanding the spectrum of germline variants in cancer
Human Genetics (2017)
-
Update of the human and mouse Fanconi anemia genes
Human Genomics (2015)
-
Genetic Counseling for Fanconi Anemia: Crosslinking Disciplines
Journal of Genetic Counseling (2014)