Abstract
Submicroscopic structural variations, including deletions, duplications, inversions and more complex rearrangements, are widespread in normal human genomes. Inverted segmental duplications or highly identical low-copy repeat (LCR) sequences can mediate the formation of inversions and more complex structural rearrangements through non-allelic homologous recombination. In a patient with 7q36 inverted duplication/terminal deletion, we demonstrated the central role of a pair of short inverted LCRs in the vasoactive intestinal peptide receptor gene (VIPR2)-LCRs in generating the rearrangement. We also revealed a relatively common VIPR2-LCR-associated inversion polymorphism disrupting the gene in almost 1% of healthy subjects, and a small number of complex duplications/triplications. In genome-wide studies of several thousand patients, a significant association of rare microduplications with variable size, all involving VIPR2, with schizophrenia was recently described, suggesting that altered vasoactive intestinal peptide signaling is likely implicated in the pathogenesis of schizophrenia. Genetic testing for VIPR2-LCR-associated inversions should be performed on available cohorts of psychiatric patients to evaluate their potential pathogenic role.
Similar content being viewed by others
Introduction
Over the past few years, there has been a strong effort in genomic research to identify submicroscopic structural variations (SVs) including deletions, duplications, insertions, inversions, translocations and more complex rearrangements in the human genome. The majority of these studies have been based on microarray analysis and focused on copy number variations (CNVs).1, 2 However, balanced rearrangements not involving gain or loss of DNA cannot readily be detected with arrays. Inversions large enough to be detectable at the resolution of cytogenetics have long been known. Shorter inversions can be globally detected by paired-end sequencing and mapping3 or by a variety of targeted methods, including pulse-field gel electrophoresis and Southern blot. Four major mechanisms account for the majority of SVs: non-allelic homologous recombination (NAHR), nonhomologous end-joining (NHEJ), fork stalling and template switching and L1-mediated retrotransposition.4 Inversions of regions located between segmental duplications or highly identical low-copy repeat (LCR) sequences may be formed by NAHR if the LCRs are in inverted orientation with respect to each other. Inverted LCRs also have a role in mediating the generation of recurrent inverted duplications/deletions,5 nonrecurrent duplications/triplications6 and more complex rearrangements.7
While performing the molecular characterization of a patient with a complex 7q36 inverted duplication/terminal deletion, we documented the involvement of inverted LCRs enclosing exons 1 and 2 of the vasoactive intestinal peptide receptor (VIPR2) gene in the genesis of the rearrangement. We also demonstrated that the same LCRs mediate the occurrence of rare duplications/triplications encompassing the whole VIPR2 gene, and of an inversion polymorphism, presumably disrupting VIPR2 expression, in almost 1% of Italian subjects.
Rare CNVs are prominently involved in the etiology of several neuropsychiatric disorders, including schizophrenia, autism and bipolar disorder.8 Two reports9, 10 recently analyzed the association of rare CNVs with schizophrenia in large cohorts of patients and control subjects. Both groups reported a significant association of rare microduplications with variable size involving the VIPR2 gene with schizophrenia. VIPR2 transcription and cAMP signaling were significantly increased in lymphocytes from duplication carriers, suggesting that altered vasoactive intestinal peptide signaling is likely implicated in the pathogenesis of schizophrenia. The molecular structure of these variations and their association to specific genomic features was not fully assessed. We present a model that explains the genesis of each of our rearrangements as well as some of the VIPR2-associated CNVs reported by Vacic et al9 and Levinson et al.10 We also suggest that decreased VIPR2 expression in inversion carriers may impact neurodevelopment and behavior; therefore, genetic testing for the inversion should be carried out in large cohorts of psychiatric patients.
Materials and methods
Patient P1 is a 5-year-old female referred to the Child Neuropsychiatry Unit at our Institute for language delay and mild mental retardation. Family history was unremarkable. She was born at 39 weeks of gestation by cesarean section for podalic presentation. Pregnancy had been uncomplicated, although fetal ultrasound had revealed intrauterine growth retardation. Birth weight was 1965, g. APGAR scores were 8 (at 1 min) and 10 (at 5 min). Motor milestones were slightly delayed: she sat at 8 months and walked at 20 months. She uttered her first words at 14 months and subsequent progression of language was delayed. At the age of 5 years, her weight was 17.5 kg (41st centile), height 98.5 cm (3rd centile) and head circumference 50.5 cm (50th centile). She showed mild mental retardation (overall IQ: 62 at Stanford-Binet and Borrell Maisonny). Language delay, speech and social difficulties were also observed. She presented mild dysmorphic features consisting in long face, saddle-nose, anteverted nares and low-set ears.
Karyotype analysis (450–550 bands) was performed on subject P1 using standard protocols. Array-CGH was performed with the Agilent oligo platform (Agilent, Santa Clara, CA, USA) at a resolution of ∼60 kb (Agilent kit 105 k). The procedures for DNA digestion, labeling and hybridization were executed according to the manufacturer’s instructions. Gender-matched genomic DNAs were obtained from individuals NA10851 (male) and NA15510 (female; Coriell Repository, Camden, NJ, USA). The quality of each DNA was evaluated by conventional absorbance measurements with a NanoDrop 1000 instrument (Thermo Scientific, Wilmington, DE, USA) and electrophoretic gel mobility assays. Quality of the experiments was assessed by using the Agilent Feature Extraction QC Metric (v10.1.1.1). The derivative log ratio spread (DLR) value was calculated with the Agilent Genomics Workbench software. Only CNVs having DLR spread values <−0.3 and >+0.3 in three or more probes were taken into consideration.
We determined VIPR2-LCR orientation on three sets of anonymous DNA samples: (a) parental samples from 324 anonymized parent/child triplets selected from a cohort of healthy Italian subjects previously collected for epidemiological studies on mental health in youth; (b) 937 randomly selected samples, among which: 243 anonymous blood donors; 263 anonymous university students; 257 anonymous clinical left-over blood samples; 174 anonymized subjects from a second cohort collected for epidemiological studies of mental health in youths; and (c) 64 EBV lines from European (20), Asian (20) and African (28) subjects obtained from the Coriell repository. The overwhelming majority of subjects in sets (a) and (b) is expected to be unrelated and of Italian ancestry.
Long-range PCRs were performed on all subjects using JumpStart Red ACCUTaq LA DNA polymerase (Sigma Aldrich, Milano, Italy) using primers F1/R1, F1/F2, F2/R2 and R1/R2. Sequencing reactions were performed with a Big Dye Terminator Cycle Sequencing kit (Applied Biosystems, Monza, Italy) and run on an ABI Prism 3130xl Genetic Analyzer (Applied Biosystems). Target sequences for quantitative PCR (qPCR) analysis were selected using the Primer Express 3.0 software (Applied Biosystems). cDNA synthesis was performed on selected Coriell lymphoblastoid lines with Ready-To-Go You-Prime First strand beads (GE HealthCare, Milano, Italy) and random hexamers. Genomic qPCR11 and VIPR2 TaqMan (Hs00173643_m1, Applied Biosystems) qRT-PCR assays were performed on an ABI PRISM 7900HT sequence detection system (Applied Biosystems).
Results
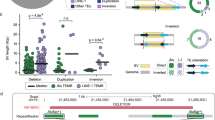
Routine karyotype analysis showed the presence of a derivative chromosome 7 with added material on the long arm, ending with fluorescent satellites: 46,XX,der(7)t(7;D or G)(7q36;p11.2; P1, Figure 1a). By array-CGH, we discovered a 14.5-Mb de novo duplication associated to a 50-kb terminal deletion of 7q (Figure 1b). No additional imbalances were present. The final interpretation was: 46,XX,der(7)t(7;D or G)(7q36; p11.2).arr 7q35 q36.3(144 063 347 × 2, 144 093 894–158 612 902 × 3, 158 747 771–158 811 268 × 1; hg18). These data suggest that the derivative chromosome was the product of an inverted duplication associated with a distal contiguous deletion (inv-dup-del)12 on which satellites from D or G chromosomes (sat D or G) were transposed. By qPCR (Figure 1c, Supplementary Table 1), we narrowed the boundaries of the duplication and demonstrated that duplicated and deleted regions were separated by a 30-kb sequence with normal copy number. This sequence is surrounded by a pair of inverted 2525 bp (chr7:158 924 495–158 927 019) and 3120 bp (chr7:158 954 809–158 957 928) LCRs with 95% identity enclosing the promoter and exons 1 and 2 of the VIPR2 gene (VIPR2-LCRs; Figure 1d, Supplementary Figure 1a). These LCRs are annotated in the GRCh37/h19 assembly of the human genome. By long-range amplification and sequence analysis of the breakpoint junction (Table 1), we demonstrated that the rearrangement is indeed an inverted duplication/deletion generated by NAHR between the VIPR2-LCRs. Both breakpoints localize to polymorphic 31-bp simple repeats (Table 1).
Cytogenetic and molecular analysis of a subject with 7q36 inverted duplication/deletion. (a) Cutout of chromosome 7 from the karyotype of subject P1 showing the elongated der 7. (b) Array-CGH plot of the whole chromosome 7 (left) and detail of 7q36 (right) showing the duplicated (green bar) and deleted (red arrow) regions. (c) qPCR analysis of the boundaries between 7q duplication and terminal deletion in P1 and her normal parents (M, mother; F, father; RQ, relative quantitation). The position of the six qPCR probes is shown in (d), partial map of the distal portion of the human VIPR2 gene, including exons 1 and 2, from the UCSC genome browser (hg19, http://genome.ucsc.edu). The relative positions of LCRs (VIPR2-REP A and B), long-range PCR primers F1, R1, F2, R2 and qPCR probes are indicated. Long-range PCRs using primers F1/F2 or R1/R2 amplify VIPR2-LCR inversions and VIPR2-LCR-associated duplications/triplications.
Inversion polymorphisms of a 4-Mb sequence enclosed between large, complex LCRs can increase the probability of recurrent pathogenic rearrangements through NAHR.13, 14, 15 Although this may not necessarily be true for rare rearrangements mediated by shorter LCRs, we analyzed VIPR2-LCR orientation by long-range PCR in the patient’s parents. Neither parent carried an inversion of the VIPR2-LCR region. We additionally analyzed 1585 healthy unrelated subjects of Italian ancestry and detected evidence of heterozygous inversion in 16 subjects (1%; Table 1). Using qPCR (Supplementary Table 1; Supplementary Figure 1a), we demonstrated that 13 of the subjects (0.82%) actually carried an inversion, whereas 3 (0.18%) had complex microduplications/triplications/quintuplications extending proximally beyond the VIPR2 gene. These rearrangements contain an inverted VIPR2-LCR segment, therefore they are also positive by long-range PCR. We further verified them by aCGH (Supplementary Figure 1b) and qPCR (Supplementary Figure 1c), and characterized the breakpoint junctions of all subjects with duplications/triplications and inversion carriers (Table 1; Supplementary Figure 1d). All junctions within the VIPR2-LCRs were generated by NAHR between the same highly polymorphic 31-bp simple repeats involved in the breakpoints of the inv dup/del 7q patient. The size of the repeats in each rearranged subject, as well as in six randomly chosen subjects without inversions or duplications, is shown in Table 1. Subject C67 carries a 274-kb duplication (chr7:158 523 272–158 770 220) and a 150-kb triplication (chr7:158 772 854–158 924 495); we were not able to clone the Jc2 junction, probably due to the presence of a large GC-rich repetitive sequence at one of the breakpoints.
Subject C190 shows a 12-kb duplication (chr7:158 788 197–158 800 731), followed by a 120-kb triplication (chr7:158 803 996–158 924 495); C182 carries triplicated and quintuplicated regions with the same boundaries as C190. Based on their size and junction sequences (Supplementary Figure 1e), C190 and C182 are likely to carry the same rearrangement in heterozygous and homozygous form, respectively. As C182 and C190 are married and their offspring also carries a duplication/triplication, we cannot exclude that C182 may carry a triplication/quintuplication on one chromosome 7. We were also unable to determine whether the two subjects are blood relatives. In either case, C182 would carry four active copies of VIPR2.
The region encompassed by the VIPR2-LCRs is duplicated in all three subjects. We observed parent-to-child transmission of both inversions (C82) and duplications (C182/C190). The inversions involve exons 1 and 2 of VIPR2 and presumably inactivate the gene (Figure 2). We also tested 68 cell lines of European, Asian and African subjects from the Coriell Repository and found inversions in NA18563 (Han Chinese) and NA07348 (European). We analyzed VIPR2 expression by qRT-PCR in the Coriell lines, but transcript levels were too low to verify decreased expression in the lines of inversion carriers compared with control lines.
Graphical representation of VIPR2-LCR-associated structural rearrangements. (a) Wild-type orientation of the VIPR2-LCRs. (b) VIPR2-LCR inversion. (c) Inverted duplication/terminal deletion in subject P1. A general mechanism for the creation of this category of rearrangements is proposed in Zuffardi et al,12 Figure 1. (d) VIPR2–LCR-associated complex duplication/triplication in subjects C62, C182 and C190. A model for the generation of this type of rearrangements is proposed in Carvalho et al,6 Figure 5. The relative positions of the VIPR2 gene and VIPR2-associated LCRs (proximal LCR, red triangle; distal LCR, yellow triangle) are indicated. Duplicated portions are shown in light blue, triplicated regions in dark blue. The position of rearrangement-specific junctions (JC1 and JC2) are also shown. Features are not in scale.
Discussion
We have analyzed the structural features underlying an inverted duplication/terminal deletion at 7q36.3 in a patient with development and language delay and mild dysmorphisms. The 14.5-Mb duplicated portion contains >80 genes, the 50-kb deletion none. Subtelomeric 7q36 deletions16 are associated with, among other pathological phenotypes, holoprosencephaly17 and Currarino syndrome,18 while duplications are rare. Novales et al19 concluded that patients with duplications covering 7q32->qter show only mild features including low birth weight, retardation of development, low-set years, small nose, skeletal anomalies, kyphoscoliosis and muscular hypotonia. Bartsch et al20 described independent small (550 kb) 7q36.3 microduplication and atypical 17q11.2 (NF1) microdeletion in a girl with neurofibromatosis and concluded that the 7q36.3 trisomy represented a subtelomeric CNV without phenotypic consequences. Lehnen et al21 reported a severe phenotype in a girl with partial tetrasomy 7q35–q36. The only 7q36 duplication reported in the Decipher database (patient 254265, http://decipher.sanger.ac.uk/) in a subject with developmental delay, prominent eyes and short stature was inherited from her normal parents. Thus, the pathogenic role of the rearrangement in patient P1 cannot be unequivocally demonstrated.
We identified the role of a pair of VIPR2-associated inverted LCRs (Figures 1d and 2a) in the genesis of the 7q-inverted duplication/deletion (Figure 2c). The same mechanism, NAHR between two large complex LCRs, generates the recurrent inverted duplication/deletion of 8p, inv dup(8p).12, 13 We also documented rare NAHR-mediated rearrangements at a pair of small inverted LCRs close to the MYOM gene on 8p.7 Most nonrecurrent inverted duplications/terminal deletions seem to be generated by NHEJ or intra-strand annealing (reviewed in Zuffardi et al12).
In addition, the VIPR2-LCRs mediate inversions of a region, enclosed between them, containing the promoter and first two exons of the VIPR2 gene22 (Figure 2b) in 13/1653 healthy subjects and cell lines. Such inversions should completely shut off VIPR2 expression on the affected chromosome, although we have not been able to verify decreased expression due to the extremely low levels of VIPR2 transcription in lymphoblastoid lines. Finally, we also detected three complex rearrangements mediated by VIPR2-LCRs, consisting of intermixed duplications and triplications in which the triplicated segment is inverted and located between directly oriented duplicated segments (Figure 2d). Two specific breakpoint junctions between duplicated and triplicated sequences (JC1 and JC2, Figure 2d) characterize all such rearrangements. In subjects C182 and C190 (Supplementary Figure 1e), the JC2 breakpoint sequences show 2-bp microhomology making the junction compatible with NHEJ or breakpoint-induced replication.
Rearrangements with the same structural organization have been found on different chromosomes in patients with a variety of physical abnormalities.6, 23 In all cases where the parental origin could be determined, the triplication was found to be composed of alleles from both homologs of one of the parents, consistent with a meiotic or pre-meiotic origin. We previously demonstrated the same arrangement in two larger nonrecurrent rearrangements and pointed out for the first time the presence of both duplicated and triplicated portions.24 To explain the genesis of complex triplications, it was suggested that intrachromosomal triplications can arise from a dicentric, inverted duplicated chromosome.25 Breakage of the dicentric chromosome and recombination with a normal chromosome would lead to triplication. Alternatively, the triplications could form by U-type exchanges between three chromatids.26, 27, 28, 29 One exchange between homologous chromatids would take place at the distal breakpoint region, the other at the proximal breakpoint region, between either sister chromatids or homologous chromosomes. Brewer et al23 proposed a replication-based mechanism, supported by yeast observations, involving the generation of a dimeric inverted circular intermediate. Unfortunately, the model fails to account for the constant presence of duplicated regions of variable, and sometimes fairly large, size in all rearrangements examined so far. Very recently, based on the molecular analysis of complex rearrangements at the MECP2 and PLP1 loci, Carvalho et al6 demonstrated the role of inverted repeats in mediating complex triplications and proposed a model for their generation. Our VIPR2-associated rearrangements perfectly fit this model.
The high-density aCGH analysis performed by Vacic et al9 illustrates in detail the variety and complexity of 7q36.6 rearrangements. Duplications and duplications/triplications of different sizes span the region with apparently random distribution. By comparing the genomic coordinates of each rearrangement, provided in Supplementary Table 3,9 with the position of the VIPR2-LCRs (chr7:158 617 161–158 650 761, in hg18), we discovered that in all samples containing complex microduplications/triplications one of the duplicated segments, adjacent to the triplicated portion, always exactly spans the VIPR2-LCR; the rearrangements are therefore compatible with Carvalho et al’s model.6 Although the size of the VIPR2-LCR-related duplication is always the same, the size and location of additional duplicated and triplicated segments is variable. Strikingly, while four of the rearrangements extend proximally from the LCR, one (00C02873) extends distally, again in agreement with Carvalho’s model.6
Although our approach was not designed to detect all VIPR2-associated CNVs, our findings complement and extend Vacic et al’s9 and Levinson et al’s10 observations. NAHR between VIPR2-associated LCRs is probably responsible for some of the rearrangements described by Vacic et al.7 In the populations examined, 7q63 rearrangements are rare (0.18% in our cohort, including the odd C182/C190 couple) or extremely rare (0.03–0.05%)9, 10 in controls and still fairly rare (0.25–0.35%)9, 10 in schizophrenia patients, whereas according to our results VIPR2-associated inversions are present in ∼0.8% of normal subjects of Italian descent. The frequency of VIPR2-associated inversions should be assessed on large cohorts of subjects of different ethnic background.
Increased, as well as decreased, VIPR2 expression may result in dysregulation of adult neurogenesis or synaptic transmission. We would then expect to find a sizeable increase in the number of inversion carriers in schizophrenic subjects or in patients with other neuropsychiatric disorders where association to VIPR2 was suspected. Alternatively, deletions and duplications may have contrasting effects on psychiatric features, as it has been demonstrated for CNVs in 1q21.1 and 16p11.2.8, 9, 10, 30 In any event, genetic testing for VIPR2 inversions, in addition to CNVs, in schizophrenia and other psychiatric diseases could potentially yield extremely useful results.
Finally, Vipr2-null mice show abnormal rest and activity rhythms;31 unless this genotype is lethal in humans, we expect to find rare (∼1:20,000 newborns) subjects carrying homozygous VIPR2 inversions and unknown, but presumably pathological, phenotype.
References
Iafrate AJ, Feuk L, Rivera MN et al: Detection of large-scale variation in the human genome. Nat Genet 2004; 36: 949–951.
Sebat J, Lakshmi B, Troge J et al: Large-scale copy number polymorphism in the human genome. Science 2004; 305: 525–528.
Tuzun E, Sharp AJ, Bailey JA et al: Fine-scale structural variation of the human genome. Nat Genet 2005; 37: 727–732.
Zhang F, Gu W, Hurles ME et al: Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet 2009; 10: 451–481.
Ciccone R, Mattina T, Giorda R et al: Inversion polymorphisms and non-contiguous terminal deletions: the cause and the (unpredicted) effect of our genome architecture. J Med Genet 2006; 43: e19.
Carvalho CM, Ramocki MB, Pehlivan D et al: Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat Genet 2011; 43: 1074–1081.
Giorda R, Ciccone R, Gimelli G et al: Two classes of low-copy repeats comediate a new recurrent rearrangement consisting of duplication at 8p23.1 and triplication at 8p23.2. Hum Mutat 2007; 28: 459–468.
Malhotra D, Sebat J : CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell 2012; 148: 1223–1241.
Vacic V, McCarthy S, Malhotra D et al: Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature 2011; 471: 499–503.
Levinson DF, Duan J, Oh S et al: Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry 2011; 168: 302–316.
Bonaglia MC, Giorda R, Mani E et al: Identification of a recurrent breakpoint within the SHANK3 gene in the 22q13.3 deletion syndrome. J Med Genet 2006; 43: 822–828.
Zuffardi O, Bonaglia M, Ciccone R et al: Inverted duplications deletions: underdiagnosed rearrangements?? Clin Genet 2009; 75: 505–513.
Giglio S, Broman KW, Matsumoto N et al: Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am J Hum Genet 2001; 68: 874–883.
Osborne LR, Li M, Pober B et al: A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat Genet 2001; 29: 321–325.
Giglio S, Calvari V, Gregato G et al: Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am J Hum Genet 2002; 71: 276–285.
Ravnan JB, Tepperberg JH, Papenhausen P et al: Subtelomere FISH analysis of 11 688 cases: an evaluation of the frequency and pattern of subtelomere rearrangements in individuals with developmental disabilities. J Med Genet 2006; 43: 478–489.
Gurrieri F, Trask BJ, van den Engh G et al: Physical mapping of the holoprosencephaly critical region on chromosome 7q36. Nat Genet 1993; 3: 247–251.
Lynch SA, Bond PM, Copp AJ et al: A gene for autosomal dominant sacral agenesis maps to the holoprosencephaly region at 7q36. Nat Genet 1995; 11: 93–95.
Novales MA, Fernandez-Novoa C, Hevia A et al: Partial trisomy for the long arm of chromosome 7. Case report and review. Hum Genet 1982; 62: 378–381.
Bartsch O, Vlcková Z, Erdogan F et al: Two independent chromosomal rearrangements, a very small (550 kb) duplication of the 7q subtelomeric region and an atypical 17q11.2 (NF1) microdeletion, in a girl with neurofibromatosis. Cytogenet Genome Res 2007; 119: 158–164.
Lehnen H, Maiwald R, Neyzen S et al: Severe phenotype in a girl with partial tetrasomy 7, karyotype 46,XX,trp(7)(q35q36). Cytogenet Genome Res 2009; 125: 248–252.
Lutz EM, Shen S, Mackay M et al: Structure of the human VIPR2 gene for vasoactive intestinal peptide receptor type 2. FEBS Lett 1999; 458: 197–203.
Brewer BJ, Payen C, Raghuraman MK et al: Origin-dependent inverted-repeat amplification: a replication-based model for generating palindromic amplicons. PLoS Genet 2011; 7: e1002016.
Giorda R, Beri S, Bonaglia MC et al: Common structural features characterize interstitial intrachromosomal Xp and 18q triplications. Am J Med Genet 2011; 155A: 2681–2687.
Schinzel AA, Brecevic L, Bernasconi F et al: Intrachromosomal triplication of 15q11–q13. J Med Genet 1994; 31: 798–803.
Wang J, Reddy KS, Wang E et al: Intrachromosomal triplication of 2q11.2–q21 in a severely malformed infant: case report and review of triplications and their possible mechanism. Am J Med Genet 1999; 82: 312–317.
Ungaro P, Christian SL, Fantes JA et al: Molecular characterisation of four cases of intrachromosomal triplication of chromosome 15q11–q14. J Med Genet 2001; 38: 26–34.
Roberts SE, Dennis NR, Browne CE et al: Characterisation of interstitial duplications and triplications of chromosome 15q11–q13. Hum Genet 2002; 110: 227–234.
Vialard F, Mignon-Ravix C, Parain D et al: Mechanism of intrachromosomal triplications 15q11–q13: a new clinical report. Am J Med Genet 2003; 118A: 229–234.
McCarthy SE, Makarov V, Kirov G et al: Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet 2009; 41: 1223–1227.
Harmar AJ, Marston HM, Shen S et al: The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell 2002; 109: 497–508.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Beri, S., Bonaglia, M. & Giorda, R. Low-copy repeats at the human VIPR2 gene predispose to recurrent and nonrecurrent rearrangements. Eur J Hum Genet 21, 757–761 (2013). https://doi.org/10.1038/ejhg.2012.235
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2012.235
Keywords
This article is cited by
-
Definition and refinement of the 7q36.3 duplication region associated with schizophrenia
Scientific Reports (2013)
-
Search for missing schizophrenia genes will require a new developmental neurogenomic perspective
Journal of Genetics (2013)