Abstract
Developmental delay/intellectual disabilities, speech disturbance, pre- and postnatal growth retardation, microcephaly, signs of ectodermal dysplasia, and genital malformations in males (hypospadias) represent the phenotypic core of the recent emerging 19q13.11 deletion syndrome. Using array-CGH for genome-wide screening we detected an interstitial deletion of chromosome band 19q13.11 in two patients exhibiting the recognizable pattern of malformations as described in other instances of this submicroscopic genomic imbalance. The deletion detected in our patients has been compared with previously reported cases leading to the refinement of the minimal overlapping region (MOR) for this microdeletion syndrome to 324 kb. This region encompasses five genes: four zinc finger (ZNF) genes belonging to the KRAB-ZNF subfamily (ZNF302, ZNF181, ZNF599, and ZNF30) and LOC400685. On the basis of our male patient 1 and on further six male cases of the literature, we also highlighted that larger 19q13.11 deletions including the Wilms tumor interacting protein (WTIP) gene, proximal to the MOR, results in hypospadias making this gene a possible candidate for this genital abnormality due to its well-known interaction with WT1. Although the mechanism underlying the phenotypic effects of copy number alterations involving KRAB-ZNF genes at 19q13.11 has not clearly been established, we suggest their haploinsufficiency as the most likely candidate for the phenotypic core of the 19q13.11 deletion syndrome. In addition, we hypothesized WTIP gene haploinsufficiency as responsible for hypospadias.
Similar content being viewed by others
Introduction
Microdeletion of 19q13.11 is an emerging syndrome mainly identified due to widespread use of genome-wide arrays. Patients share some common and indeed distinctive findings, and this deletion could potentially represent a clinically identifiable syndrome mainly characterized by developmental delay/intellectual disabilities (DD/ID), speech disturbance, pre- and postnatal growth retardation, microcephaly, signs of ectodermal dysplasia, and genital malformations in males (hypospadias). By using array-CGH for genome-wide screening, we have detected an interstitial deletion of chromosome 19q13.11 in two patients exhibiting the recognizable pattern of malformations as described in other instances of this cryptic genomic imbalance.1, 2, 3 The deletion detected in our patients has been compared with the five previously published cases and two reported in the Database of Chromosomal Imbalance and Phenotype in Humans using Ensemble Resources (DECIPHER), leading to the refinement of the critical region for this microdeletion syndrome to 324 kb.
Patients and methods
Clinical report
Patient 1
This patient, a boy, was the only child born to non-consanguineous healthy parents. Family history was unremarkable. During pregnancy, fetal ultrasound scans at 30 weeks of gestation revealed intrauterine growth retardation and a ventricular septal defect. Cesarean section was performed at 34 weeks of gestation, due to fetal distress. Birth weight was 1590 g (<3rd centile). Apgar scores were 8 at both 1 and 5 min. Some minor dysmorphic facial features, abnormalities of extremities (second and third finger syndactyly of the right hand and bilateral two to four partial syndactyly of toes), and proximal hypospadias were observed at birth. The early infancy was compromised by feeding difficulties and failure to thrive. At 2 years of age, a partial growth hormone (GH) deficiency was diagnosed (GH peak of 7.23 ng/ml after glucagon and of 2.01 ng/ml after arginine, normal value >10 ng/ml), which required GH treatment. Delayed milestones were noticed in the first months of life (free walking at 18 months). From the age of 4 he presented abnormal dystonic postures of the lower limbs. The dystonia, involving initially only the lower limbs, progressed to generalized dystonia, complicated by tremor, and non-epileptic myoclonus, over the following years. Generalized and oromandibular dystonia did not respond, or only partially responded, to oral antidystonic treatment (particularly, pimozide was withdrawn due to long Q–T interval after a few days of treatment), botulinum toxin treatment, and intrathecal baclofen.
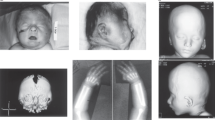
At 14 years of age, on physical examination, head circumference was 47 cm (< 2nd centile), height 124 cm (<3rd centile), and weight 27 kg (<3rd centile). He presented severe ID with absent speech and severe dystonic tetraplegia, resulting in bilateral ankle deformity, bilateral hip luxation, and multiple muscle contractures (Figure 1a). Continuous spasmodic oromandibular dystonia caused life-threatening breathing difficulties (Figure 1b). Dysmorphologic evaluation revealed: narrow face, thick medially sparse eyebrows, hypertelorism, columella below alae nasi, thin lips, retrognathia, low set ears with large lobules (Figure 1b). In addition, cutis aplasia in midline of scalp was noted (Figure 1c). Both hands had long fingers and presented bilateral clinodactyly of the fifth finger. Moreover, second and third finger syndactyly of the right hand and bilateral two to four partial syndactyly of toes were present.
Patient 1 at 14 years of age showing dystonic tetraplegia (a) with severe oromandibular impairment (b), thin face, thick eyebrows medially sparse, hypertelorism, columella below alae nasi, thin lips, low set ears with large lobules (b), and cutis aplasia in midline of scalp (c). Patient 2 at 8 years of age showing round face with narrow and up-slanting palpebral fissures, thick eyebrows medially sparse, puffy checks, columella below alae nasi, short philtrum, thin lips, hypodontia with coarse and rudimental shaped teeth (d), small mandible, and large ear lobules (e).
The laboratory examinations including routine blood and urine tests were normal. Brain magnetic resonance imaging (MRI) revealed mild enlargement of the lateral ventricles. Karyotype, subtelomeric fluorescence in situ hybridization (FISH), FISH for the DiGeorge syndrome critical region, and molecular investigations for dystonia (mutations of the DYT1 gene) gave normal results.
Patient 2
This patient, a girl, was the third born to unrelated healthy parents. She had two older healthy siblings, a brother and a sister, respectively. There was no history of hereditary disorders in her family. Two relatives, second-degree and third-degree, respectively, had a history of learning disabilities. The pregnancy was complicated by intrauterine growth retardation, whereas the spontaneous vaginal delivery, at 38 weeks of gestation, was uncomplicated. Birth weight was 1.9 kg (<3rd centile). The neonatal and postnatal periods were uneventful. A sacral dimple was noted at birth, however, no occult spinal dysraphism was identified through spine MRI. The early infancy was compromised by failure to thrive without evident feeding difficulties. She presented moderate global DD with motor and verbal domains predominantly impaired.
At 8 years of age, on physical examination, head circumference was 48 cm (< 2nd centile), height 120 cm (<3rd centile), and weight 27 kg (50th centile). She presented moderate ID with impaired expressive language, partially due to abnormal tongue movement consistent with oral dyspraxia, and hoarse voice. Neurological examination revealed poor balance with clumsiness, dysdiadochokinesia and mild hypotonia. She wore glasses for astigmatism. Dysmorphologic evaluation revealed: round face with narrow and up-slanting palpebral fissures, thick medially sparse eyebrows, puffy cheeks, columella below alae nasi, short philtrum, thin lips, hypodontia with coarse and rudimental shaped teeth, small mandible, large ear lobules, long fingers with short fifth digits bilaterally (Figures 1d and e). No other abnormalities were detected on clinical examination.
Array-CGH
Array-CGH was performed on patient 1 and his parents using the Agilent kit 4x180K (Human Genome CGH Microarray, Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer's protocol. For patient 2, array-CGH was performed using a custom-designed 4x180K Agilent array V8.1, and data were analyzed as described previously.4, 5
CNVs reported in the Database of Genomic Variants http://projects.tcag.ca/variation/ and in-house databases of benign CNVs were excluded from further analysis. Genomic positions are referred to the Human Genome March 2006 assembly (hg18).
Fluorescence in situ hybridization
FISH was performed on patients’ parents by using BAC RP11-630G5 (patient 1), and RP11-770P14 (patient 2), mapping within the deleted regions.
Results
Array-CGH analysis identified a 1.7 Mb deletion at 19q13.11-q13.12 in patient 1 (proximal breakpoint located between 39 608 712 and 39 626 575 bp, distal breakpoint located between 41 330 875 and 41 353 676 bp), and an ∼2.63-Mb interstitial deletion at 19q13.11-q13.12 in patient 2 (proximal breakpoint located between 39 735 455 and 39 803 651 bp, distal breakpoint located between 42 436 832 and 42 595 049 bp; Figures 2a and b). The two informative microsatellites (D19S425 and D19S893) showed that the deletion in patient 1 occurred on the maternal chromosome 19 (Figure 2c).
Parental FISH analysis confirmed that both deletions were de novo. Possible insertional translocations were also excluded in all parental samples (data not shown).
Discussion
We identified two unrelated individuals harboring overlapping de novo deletions of the long arm of chromosome 19 of about 1.7 and 2.6 Mb, respectively.
Five cases with 19q13.11 deletion have been published1, 2, 3 with two additional ones present in the DECIPHER database (patients 127 and 3776). Only one of these patients had a cytogenetically visible deletion.3 Malan et al, describing the first three patients, indicated the recognizable clinical pattern of this chromosome imbalance and Schuurs-Hoeijmakers et al comparing the clinical features of 19q13.11 deletion cases narrowed the critical region to 750 kb.1, 2 Typical features, that were also present in both of our patients, are DD/ID, speech disturbance, microcephaly, pre- and postnatal growth retardation, and some signs of ectodermal dysplasia represented in our cases by cutis aplasia in midline of scalp and sparse eyebrows in patient 1 and by sparse eyebrows and teeth anomalies in patient 2. Consistent with the notion that male genital abnormality is one of the important recognizable characteristics of this syndrome, hypospadias is observed in all the male patients including patient 1, the four previously published ones and the two DECIPHER database cases. In addition, all the patients present some facial dysmorphisms, although a peculiar facial phenotype cannot be defined. Extremity anomalies (mainly clinodactyly of the fifth finger and syndactyly) are common but not necessarily present in all individuals. Table 1 summarizes clinical findings of our patients and of the previously published ones.
The deletion reported in all the previous cases overlapped with the deletions in our patients, leading to a further narrowing of the minimal overlapping critical region to a size of 324 kb, located between nucleotide position 39 803 651 and 40 127 916 on chromosome 19q (UCSC Genome Browser, Build 36.1, hg18; Figure 3). Hence the minimal overlapping region (MOR) includes five genes, the ZNF302, ZNF181, ZNF599, LOC400685, and ZNF30. Predictions of candidate genes are difficult as the function of some of them is still largely unknown. Schuurs-Hoeijmakers et al had considered potential candidate genes LSM14A, UBA2, and, to a lesser extent, the four zinc finger (ZNF) genes belonging to the KRAB-ZNF subfamily (ZNF302, ZNF181, ZNF599, and ZNF30).2 The first two genes are not included in deleted regions in our cases. These KRAB-ZNF genes might be considered the strongest candidate for 19q13.11 microdeletion syndrome, though, to date, the role of haploinsufficiency for these genes in human diseases is not clearly established. The KRAB-ZNF genes constitute the largest single family of transcriptional regulators in mammals and are expressed in all human tissues being involved in the embryonic development, cell differentiation, cell proliferation, apoptosis, neoplastic transformation, and cell cycle regulation.6, 7 However, despite their prevalence, most KRAB-ZNF genes remain completely uncharacterized in their function.
A schematic representation of the overlapping deletions in the two patients and previously reported cases. A close view of chromosome band 19q12-q13.12 is displayed on the top. The comparison of the deleted regions in our patients and previously reported cases narrows down the overlapping critical region from about 750 kb to a size of 324 kb, located on chromosome band 19q13.11, encompassing five genes. WTIP (position: chr19:39 664 720–39 683 925) and SCN1B (position: chr19:40,213,432–40,217,014) are located outside the MOR (chr19: 39 803651–40 127 916).
All the reported male patients presented hypospadias and shared the deletion of Wilms tumor interacting protein (WTIP), that is located outside from the MOR common to all cases (Figure 3). This gene is indeed a strong candidate for the observed sexual development disorder as its product interacts with WT1 (Wilms tumor 1), a gene having a well-known role in proper sex development.8 The Wt1 protein has an essential role in the mammalian urogenital development, and WT1 mutations in humans lead to a variety of disorders, including Wilms’ tumor, as well as Frasier and Denys–Drash syndromes. Phenotypic anomalies in the latter two syndromes include pseudohermaphroditism and sex reversal in extreme cases.9 Although mutations for WTIP have never been reported, its well-demonstrated interaction with WT1 makes it the best candidate in the 19q13.11 region for the observed hypospadias.10, 11 Moreover, considering that patient 127 in the DECIPHER database, also with a deletion involving WTIP, has an XX karyotype with cryptorchid testes, we might even hypothesize that, as most genes whose mutations are associated with sex developmental disorders,12, 13, 14 WTIP haploinsufficiency may result in a range of sex disorders starting from the first stage of gonadal differentiation (XX males) to the later stages in which urethral fusion occurs (XY males with hypospadias). This is consistent with the WT1 expression pathway both in the bipotential genital ridge and subsequently in the differentiated gonads.11, 15
Comparing all the reported patients, we confirm a significantly more severe phenotype in patient 1 than in others concerning the presence of a disabling movement disorder and the severity of DD/ID. We did not identify any genes in the deleted region that can currently be considered as an obvious candidate for dystonia that dominates the clinical phenotype of our patient 1. Therefore, a synergistic effect between several genes may be a predisposing factor for this phenotypical feature and, eventually, other genetic, epigenetic, or environmental factors would be clearly required for the full expression of dystonia, according to the second hit model.16 Alternatively, changed expression/regulation of genes in the vicinity of the deletion might contribute to the more severe phenotype of this patient, or, finally, the deletion might unmask a recessive mutation in a gene responsible for dystonia.17 In addition, patient 1 had a long Q–T episode related to pimozide treatment. It is well known that neuroleptics can be associated with increased risk of cardiac arrhythmias, including prolongation of Q–T interval, and sudden death due to inhibition of cardiovascular Na+, Ca2+, and K+ channels.18 The 19q13.11 deleted region includes the SCN1B gene encoding a sodium channel β-1 subunit whose heterozygous mutations may result in generalized epilepsy with febrile seizures plus, Brugada syndrome 5, and defects in cardiac conduction.19, 20 Thus, SCN1B haploinsufficiency might have a synergistic effect with pimozide making manifest an otherwise hidden abnormal repolarization defect. Consequently, this observation might suggest that specific care in using neuroleptics should be indicated for patients with heterozygous null mutations in voltage-gated sodium channels. Considering all the published patients with microdeletion of 19q13.11, 6 out 7 (Table 1) harbored the deletion of SCN1B, however, none suffered from epilepsy or cardiac conduction defects, at least in pediatric age, in agreement with the finding that even deleterious ion channel mutations confer uncertain risk to an individual depending on the other variants with which they are combined.21
In conclusion, we have identified two patients with a deletion in chromosome band 19q13 that allowed us to further narrow down the critical region for 19q13.11 deletions to 324 kb. This region encompasses five genes. We suggest that KRAB-ZNF genes located within the deleted region are the most likely candidate genes for the phenotypic core of the 19q13.11 deletion syndrome, consistent with their importance for the embryonic development and cell functions. Second, we hypothesized WTIP haploinsufficiency as responsible for hypospadias or, to a lesser extent, for 46,XX sex reversal.
References
Malan V, Raoul O, Firth HV et al: 19q13.11 deletion syndrome: a novel clinically recognisable genetic condition identified by array comparative genomic hybridisation. J Med Genet 2009; 46: 635–640.
Schuurs-Hoeijmakers JH, Vermeer S, van Bon BW et al: Refining the critical region of the novel 19q13.11 microdeletion syndrome to 750 Kb. J Med Genet 2009; 46: 421–423.
Kulharya AS, Michaelis RC, Norris KS, Taylor HA, Garcia-Heras J : Constitutional del(19)(q12q13.1) in a three-year-old girl with severe phenotypic abnormalities affecting multiple organ systems. Am J Med Genet 1998; 77: 391–394.
Bonaglia MC, Giorda R, Beri S et al: Molecular mechanisms generating and stabilizing terminal 22q13 deletions in 44 subjects with Phelan/McDermid Syndrome. PLoS Genet 2011; 7: e1002173.
Boone PM, Bacino CA, Shaw CA et al: Detection of clinically relevant exonic copy-number changes by array CGH. Hum Mutat 2010; 31: 1326–1342.
Frietze S, O’Geen H, Blahnik KR, Jin VX, Farnham PJ : ZNF274 recruits the histone methyltransferase SETDB1 to the 3′ ends of ZNF genes. PLoS One 2010; 5: e15082.
Urrutia R : KRAB-containing zinc-finger repressor proteins. Genome Biol 2003; 4: 231.
Srichai MB, Konieczkowski M, Padiyar A et al: A WT1 co-regulator controls podocyte phenotype by shuttling between adhesion structures and nucleus. J Biol Chem 2004; 279: 14398–14408.
Regev M, Kirk R, Mashevich M, Bistritzer Z, Reish O : Vertical transmission of a mutation in exon 1 of the WT1 gene: lessons for genetic counseling. Am J Med Genet A 2008; 146A: 2332–2336.
Wilhelm D, Palmer S, Koopman P : Sex determination and gonadal development in mammals. Physiol Rev 2007; 87: 1–28.
Pelletier J, Schalling M, Buckler A et al: Expression of the Wilms’ tumor gene wt-1 in the murine urogenital system. Genes Dev 1991; 5: 1345–1356.
Vetro A, Ciccone R, Giorda R et al: XX males SRY negative: a confirmed cause of infertility. J Med Genet 2011; 48: 710–712.
Benko S, Gordon CT, Mallet D et al: Disruption of a long distance regulatory region upstream of SOX9 in isolated disorders of sex development. J Med Genet 2011; 48: 825–830.
Allali S, Muller JB, Brauner R et al: Mutation analysis of NR5A1 encoding steroidogenic factor 1 in 77 patients with 46, XY disorders of sex development (DSD) including hypospadias. PLoS One 2011; 6: e24117. Epub 2011 Oct 20.
Morrison AA, Viney RL, Saleem MA, Ladomery MR : New insights into the function of the Wilms tumor suppressor gene WT1 in podocytes. Am J Physiol Renal Physiol 2008; 295: 12–17.
Girirajan S, Rosenfeld JA, Cooper GM et al: A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet 2010; 42: 203–209.
Lesnik Oberstein SA, Kriek M, White SJ et al: Peters Plus syndrome is caused by mutations in B3GALTL, a putative glycosyltransferase. Am J Hum Genet 2006; 79: 562–566.
Pacher P, Kecskemeti V : Cardiovascular side effects of new antidepressants and antipsychotics: new drugs, old concerns? Curr Pharm Des 2004; 10: 2463–2475.
Scheffer IE, Berkovic SF : Generalized epilepsy with febrile seizures plus: a genetic disorder with heterogeneous clinical phenotypes. Brain 1998; 120: 479–490.
Watanabe H, Koopmann TT, Le Scouarnec S et al: Sodium channel beta-1 subunit mutations associated with Brugada syndrome and cardiac conduction disease in humans. J Clin Invest 2008; 118: 2260–2268.
Klassen T, Davis C, Goldman A et al: Exome sequencing of ion channel genes reveals complex profiles confounding personal risk assessment in epilepsy. Cell 2011; 145: 1036–1048.
Acknowledgements
We thank the patients and their families for contributing to this research study. This work was supported by the Ricerca Corrente RC11017C, Fondazione IRCCS ‘C Mondino’, Pavia, to PV.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Gana, S., Veggiotti, P., Sciacca, G. et al. 19q13.11 cryptic deletion: description of two new cases and indication for a role of WTIP haploinsufficiency in hypospadias. Eur J Hum Genet 20, 852–856 (2012). https://doi.org/10.1038/ejhg.2012.19
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2012.19
Keywords
This article is cited by
-
WTIP upregulates FOXO3a and induces apoptosis through PUMA in acute myeloid leukemia
Cell Death & Disease (2021)
-
Failure to thrive - an overlooked manifestation of KMT2B-related dystonia: a case presentation
BMC Neurology (2020)
-
Cystinuria in a patient with 19q12q13.1 deletion
CEN Case Reports (2016)
-
TSHZ3 deletion causes an autism syndrome and defects in cortical projection neurons
Nature Genetics (2016)
-
Cutis Aplasia as a clinical hallmark for the syndrome associated with 19q13.11 deletion: the possible role for UBA2 gene
Molecular Cytogenetics (2015)