Abstract
Osteoporosis-pseudoglioma sydrome (OPPG) is an autosomal recessive disorder with early-onset severe osteoporosis and blindness, caused by biallelic loss-of-function mutations in the low-density lipoprotein receptor-related protein 5 (LRP5) gene. Heterozygous carriers exhibit a milder bone phenotype. Only a few splice mutations in LRP5 have been published. We present clinical and genetic data for four patients with novel LRP5 mutations, three of which affect splicing. Patients were evaluated clinically and by radiography and bone densitometry. Genetic screening of LRP5 was performed on the basis of the clinical diagnosis of OPPG. Splice aberrances were confirmed by cDNA sequencing or exon trapping. The effect of one splice mutation on LRP5 protein function was studied. A novel splice-site mutation c.1584+4A>T abolished the donor splice site of exon 7 and activated a cryptic splice site, which led to an in-frame insertion of 21 amino acids (p.E528_V529ins21). Functional studies revealed severely impaired signal transduction presumably caused by defective intracellular transport of the mutated receptor. Exon trapping was used on two samples to confirm that splice-site mutations c.4112-2A>G and c.1015+1G>T caused splicing-out of exons 20 and 5, respectively. One patient carried a homozygous deletion of exon 4 causing the loss of exons 4 and 5, as demonstrated by cDNA analysis. Our results broaden the spectrum of mutations in LRP5 and provide the first functional data on splice aberrations.
Similar content being viewed by others
Introduction
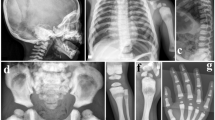
Osteoporosis-pseudoglioma syndrome (OPPG; OMIM #259770) is a rare autosomal recessive disorder characterized by early-onset severe osteoporosis and blindness.1, 2, 3, 4, 5 The phenotype is variable even among siblings.3 Osteoporosis usually manifests in early childhood with recurrent long bone fractures, vertebral compression fractures and reduced bone mineral density (BMD). More severely affected patients may have muscle weakness, bowing of the long bones and severe spinal deformities.3, 4, 5, 6 The eye phenotype is secondary to defective vascularization and ranges from congenital phthisis bulbi to milder vitreoretinal changes. Most patients are congenitally blind, or become blind in early childhood, and all are blind by the age of 25 years.1, 2, 3 Usually mental development is normal but ∼25% have cognitive impairment.1 To date, over 60 patients have been reported worldwide.
In 2001, mutations in the gene encoding low-density lipoprotein receptor-related protein 5, LRP5, were detected in patients with OPPG. No other genes are known to be associated with the phenotype.1, 7 Numerous loss-of-function mutations have been shown to cause OPPG in individuals with homozygous or compound heterozygous mutations.1, 7, 8, 9, 10, 11 Heterozygous carriers of LRP5 mutations have reduced BMD, suggesting a dominant negative effect on bone mass.2, 12 Mutations in LRP5 have also been linked to the recessive form of familial exudative vitreoretinopathy (FEVR).13Gain-of-function mutations in LRP5 can cause autosomal dominant disorders with increased bone mass.14, 15, 16
The LRP5 gene consists of 23 coding exons and is expressed in many tissues. LRP5 functions as a transmembrane coreceptor in the canonical Wnt (wingless) signaling pathway, which regulates growth and differentiation of osteoblasts.7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 In addition to direct effects on osteoblast function, LRP5 may function indirectly by inhibiting the expression of Tph1, the rate-limiting biosynthetic enzyme for serotonin in the duodenum. Inhibition of Tph1 results in augmented blood levels of serotonin, leading to inhibition of osteoblast proliferation and reduced bone formation.18
Of the ∼60 hitherto-published LRP5 mutations associated with OPPG or FEVR, only four are splice-site mutations.1, 13, 19 We present clinical and molecular data on four additional individuals with OPPG and new mutations; three mutations affecting LRP5 splicing and one deletion.
Materials and methods
Clinical and imaging data
This study was performed with the permission of Research Ethics Committees of Helsinki University Hospital, Helsinki, Finland; Hospital for Sick Children, Toronto, Canada; and University Hospital of Cologne, Cologne, Germany. Written informed consent was obtained from the patient and/or the parents. Clinical data were collected from hospital records. BMD was measured with dual-energy X-ray absorptiometry (DXA) (Lunar Prodigy, GE Lunar, Madison, WI, USA); results are presented in Z-scores, that is, in standard deviation units relative to mean values for equipment-specific age- and sex-matched reference population. Vertebral morphology was assessed by spinal radiography.
Genetic studies
DNA (patients 1–4) and RNA (patients 3–4) studies
Genomic DNA was extracted from peripheral blood samples using standard procedures. PCR amplification products of all 23 coding exons and exon–intron boundaries of LRP5 were analyzed by direct sequencing. Primers for standard PCR and sequencing were designed from genomic sequence (NM_002335), using the web-based primer program Primer3 (http://frodo.wi.mit.edu/).
RNA was extracted from peripheral blood (patient 3) or from fibroblasts (patient 4). Peripheral blood RNA was isolated with a PAXgene Blood RNA Kit (PreAnalytix, Hombrechtikon, Switzerland). RT-PCR was performed with primers located in exons 5 and 9 (forward: 5′-GTGTGCAGCTGCAGGACAAC-3′ and reverse: 5′-TGGTCTCGAGGGAGATCCTG-3′) by using the OneStep RT-PCR Kit (Qiagen, Hilden, Germany) and the amplicons were sequenced. In silico splice-site prediction was performed with http://www.fruitfly.org/seq_tools/splice.html. RNA from cultured confluent fibroblasts was extracted with Versagene RNA Cell Kit (Gentra Systems, Minneapolis, MN, USA) after 4-h incubation in a medium containing 10 μg/ml cycloheximide (CHX) to prevent nonsense-mediated RNA decay.20 The cDNA was amplified by PCR with primers located in exons 3 and 6 (forward: 5′-CCGGAAGATCATTGTGGACT-3′; reverse 5′-CGTGTCGGTCCAGTAGAGGT-3′), and sequenced.
The sequence changes observed in the patients were excluded in 171 Finnish and 96 CEPH control samples by direct sequencing. The nucleotide change in patient 3 was also excluded in 50 Turkish control samples by digestion of a PCR fragment with the restriction enzyme Hphl. The cDNA sequencing (patient 4) was performed in similarly cultured, CHX-incubated, purified and amplified fibroblast control samples.
Splicing assay (patients 1 and 2)
Appropriate tissue samples for RNA extraction were unavailable for patients 1 and 2. Consequences of their mutations were analyzed using an exon trapping assay, based on modified Exon Trapping System (Gibco BRL, Gaithersburg, MD, USA).21 Genomic DNA samples spanning exon 20 (patient 1) and exon 5 (patient 2) and adjoining introns were PCR amplified and PCR products were used for the assay. The products were cloned into the PCR 2.1-TOPO Vector (Invitrogen, Carlsbad, CA, USA). Eco-RI restriction endonuclease was used for further subcloning of the fragments into the exon-trapping vector, pSPL3 (Gibco BRL). The pSPL3 vector with inserts was then sequenced to identify the constructs that contained the wild-type sequence and the mutant sequence, and to confirm their orientation. The pSPL3 clones were transfected by lipofection into COS-7 cells.21 The transfected cells were incubated for a further 24 h at 37°C in a 5% CO2 incubator. Total RNA was then isolated using an RNAeasy Mini Kit (Qiagen), followed by synthesis of cDNA (Invitrogen). PCR was carried out for 40 cycles, using primers SD6 and SA2 that flanked the vector intron containing the inserted fragments with exons 20 and 5.21 The RT-PCR products were resolved on 1.2% agarose gels and stained with ethidium bromide.
Expression and signal-transduction studies (patient 3)
The generation of the following constructs has been described previously:1, 25 full-length human LRP5 (LRP5-WT9L) and a truncated form of human LRP5 (LRP5N-WT9L); mutant LRP5 with a deletion of six out of nine consecutive leucine residues in the signal peptide caused by the c.43_60del mutation (LRP5-Mut3L) and its truncated form (LRP5N-Mut3L). All these constructs have a myc epitope tag at the carboxy terminus. The LRP5N constructs are truncated after Pro1389 in exon 20 and lack the transmembrane and cytoplasmic domains. Other expression vectors used for Wnt signaling assays in this study are the Firefly luciferase Topflash TCF reporter plasmid (Upstate Biotechnology, Lake Placid, NY, USA) for detecting canonical Wnt signaling activity,22 Renilla luciferase pRL-TK (Upstate Biotechnology), pcDNA3.1-LacZ (Invitrogen), plasmids encoding Norrin and Frizzled-4 (FZD4),23 and the plasmids MesdC2 and RAP.24 These plasmids have been described previously.1, 25
The identified p.E528_V529ins21 mutation was amplified by OneStep RT-PCR (Qiagen) from patient RNA (forward primer: 5′-GTGTGCAGCTGCAGGACAAC-3′ and reverse primer: 5′- GGTAGTCAAGGCCAAACTCC-3′) and introduced as a 1.1-kb PshAI (NEB) restriction fragment into full-length LRP5-WT9L and LRP5N-WT9L, respectively. The dual luciferase reporter assay for Wnt/Norrin signal transduction, secretion assay and western blot analyses were performed as described previously.1, 25 Western blot analyses of conditioned medium and of cell lysates from transfected cells were carried out using an anti-myc antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for immunodetection of LRP5. An anti-β-actin antibody (Sigma, St. Louis, MS, USA) to stain β-actin served as a loading control for equal amounts of cell lysates. SDS-PAGE was performed under reducing conditions with 3–8% gradient gels. The dual luciferase reporter assay was performed as follows: HEK293T cells were transiently transfected with LRP5 (LRP5-WT9L, LRP5-p.E528_V529ins21 or LRP5-Mut3L), Norrin, FZD4, Topflash, and Renilla. Firefly luciferase activity was measured and normalized to Renilla luciferase activity (to control for transfection efficiency) 30 h after transfection. The experiment was performed four times, with each transfection performed in triplicate. The mean luciferase activity for each triplicate was calculated. The separation of membrane and cytosolic proteins was carried out by ultracentrifugation, as described elsewhere.26 Cytosolic and membrane fractions were analyzed by western blotting following reducing SDS-PAGE.25 Endogenous β-actin and calnexin, which were detected with an anti-β-actin antibody (SIGMA) and an anti-calnexin antibody (BD Biosciences, Franklin Lakes, NJ, USA), served as positive controls for proper separation of cytosolic and membrane proteins. The truncated constructs (LRP5N-WT9L, LRP5N-p.E528_V529ins21, LRP5N-Mut3L) were only used in the secretion assay. They are secreted into the medium if trafficked normally.
Results
Clinical findings
Patient 1 is a 13-year-old girl with non-consanguineous parents of Eastern European origin. Her psychomotor development is normal. She was congenitally blind and was diagnosed with exudative retinopathy at 1 month of age. She sustained a lower-leg fracture at the age of 2 years and a forearm fracture at 5 years of age. At the age of 5 years and 10 months, she had acute back pain and was diagnosed with multiple vertebral compression fractures and severe osteoporosis (lumbar spine BMD Z-score −3.8). A clinical diagnosis of OPPG was made and bisphosphonate treatment was administered from 6 to 10 years of age. She received intravenous dinatriumpamidronate (Aredia, Novartis, Basel, Switzerland) at a dose of 1.0 mg/kg/day for three consecutive days every 4 months. Her lumbar spine BMD Z-score increased to −1.6 and spinal compression fractures improved with treatment. Follow-up of laboratory tests was typical for bisphosphonate treatment; bone turnover markers decreased and calcium/PTH homeostasis remained normal. Her parents reported improved physical well being and increased strength with treatment. Her BMD and clinical status have remained stable after discontinuation of bisphosphonate treatment. Both parents are osteopenic (lumbar BMD Z-score -2.1) and their vision and eye examinations are normal. The mother has sustained one forearm fracture.
Patient 2 is an 11-year-old significantly mentally retarded boy, the only child of consanguineous parents from Iran. Between the age of 1 ½ and 2 years, the boy’s developmental progress came to an abrupt halt and his progress has since been slow. He has complex partial seizures, which have been well controlled with anticonvulsant medication since the age of 9 years. His brain MRI is normal. He is congenitally blind with bilateral retrolental masses. He sustained four femoral fractures between 3 and 7 years of age, and has not been able to walk independently after the age of 7 years. At 10 years of age, his lumbar spine BMD Z-score was −2.0 and the total body BMD Z-score was −3.3. Upon examination he was found to be hypotonic. He was stable when sitting and could stand with support. At the age of 10.5 years, bisphosphonate therapy was commenced (alendronate 70 mg, orally once a week). At the age of 11 years, the lumbar spine BMD Z-score had increased to −1.8 and total body BMD Z-score to −2.7. Both parents have osteopenia, use glasses for reading but report no other visual problems. The father has adult-onset insulin-dependent diabetes.
Patients 3A and 3B are 33- and 30-year-old brothers of Turkish origin, whose parents are first cousins. Both are of normal intelligence and were diagnosed with congenital blindness due to retrolental fibroplasia. They sustained multiple fractures (radius, humerus, and/or the hip) from minimal trauma (from age 11 and 7 years, respectively), resulting in skeletal deformities and immobility. They have been wheelchair-dependent from the age of 16 and 9 years, respectively. BMD of patient 3A revealed severe osteoporosis; his current lumbar spine BMD Z-score is −5.3. The father (age 55) has osteoporosis (lumbar BMD Z-score −3.1) and the mother (age 55) is osteopenic with a BMD Z-score of −1.2.
Patient 4 is a 17-year-old son of Tunisian parents who are first cousins. The family history is positive for OPPG in a brother and two first cousins, also sons of consanguineous parents. Eleven additional siblings of patient 4 are reportedly unaffected. No additional family members were available for this study. Psychomotor development and brain MRI are normal. The patient is blind; his visual problems were noted at 10 months of age and the ophtalmologic findings included bilateral cataract, right microphthalmia, left retinal calcifications and absent visual evoked potentials. At 6 years of age, left tibial bowing was noted. Right lower limb and spinal deformities developed subsequently. The patient has not had limb fractures. The patient can stand unsupported but walks with support mainly because of visual problems. A recent radiographic skeletal survey revealed generalized osteoporosis, scoliosis, dorsal kyphosis, severe flattening and biconcave appearance of the vertebrae, and bowing and deformation of the long bones of the lower limbs, with a ‘serpentine’ left fibula.
LRP5 mutation and splicing analysis
LRP5 screening revealed that patient 1 is a compound heterozygote for a splice-site mutation c.4112-2A>G (intron 19) and a missense mutation c.1067C>T (S356 L) in exon 6, which has been previously reported.1 The patient's mother is heterozygous for the c.1067C>T and her father for the c.4112-2A>G mutation. A heterologous splicing assay was undertaken to confirm that the latter mutation impaired splicing (Figure 1). The pSPL3 plasmid used in the heterologous splicing assay contains splice donor and acceptor sites that flank an intron containing a multiple cloning site into which fragments were inserted.21 In the absence of an inserted fragment, transfected COS-7 cells removed the endogenous intron to generate a vector/vector product of 263 bp (Figure 1a, lane 6; Figure 1b, (β splice product)). In the presence of an inserted wild-type fragment, two products including a vector/vector product of 263 bp and a correctly spliced vector/genomic product were produced (Figure 1, lane 2). Mutant constructs did not produce any detectable vector/genomic product from the mutant insert obtained from patient 1 (Figure 1a, lane 3). The substitution of the splice acceptor site (AG to GG) of intron 19 promotes splicing-out of exon 20 and flanking introns generating a vector/vector spliced product.
Splicing assay with the exon trapping assay. (a) Electrophoresis of cDNA-PCR products generated from the wild-type and mutant constructs (exon 20 from patient 1 and exon 5 from patient 2) after transfection into COS-7 cells. Lane 1: DNA marker, GeneRuler DNA Ladder Mix (MBI Fermentas, Glen Burnie, MD, USA); lane 2: splicing product of exon 20 wild-type construct; lane 3: splicing product of exon 20 mutant construct; lane 4: splicing product of exon 5 wild-type construct; lane 5: splicing product of exon 5 mutant construct; lane 6: splicing product of pSPL3. In the absence of an inserted fragment, a vector/vector splice product of 263 bp is produced (A: lane 6, B: β splice product). In the presence of an inserted wild-type fragment, splicing of the vector gives rise to two products: a vector/vector product of 263 bp and a correctly spliced vector/genomic product containing the inserted exon (237 or 132 bp; A: lanes 2 and 4, B: α). The abundance of vector/genome product as compared with the vector/vector product indicates that no enhancement of exon 20- or 5-skipping occurred in the wild-type constructs. Mutant constructs did not produce any detectable vector/genomic product from the mutant inserts obtained from patients 1 and 2 (lanes 3 and 5), indicating complete splicing-out of exons 20 or 5. (b) Genomic DNA constructs showing the cloned sequence within the exon-trapping vector, pSPL3. The sizes of vector DNA (white box), intronic DNA (lines) and exonic DNA (black box with white dots) are indicated. Two possible splicing pathways, α (vector/genome) and β (vector/vector) are shown with their respective spliced products.
Patient 2 carries a homozygous splice-site mutation c.1015+1G>T (intron 5), and both parents are confirmed to be heterozygous for the mutation. The heterologous splicing assay of this mutant construct, containing a substitution in the donor site from GT to TT, showed that exon 5 with its adjoining introns is consistently spliced out as illustrated by the 263-bp vector/vector product (Figure 1a, lane 5), as opposed to the presence of a 395-bp vector/genomic product produced by the vectors containing the wild-type exon 5 fragment (Figure 1a, lane 4).
Patients 3A and 3B were found to be homozygous for c.1584+4A>T in intron 7 of LRP5. The mutation was present in the heterozygous state in the patients’ parents and was absent on 100 chromosomes from healthy Turkish controls. The wild-type splice donor site is predicted in silico to be spliced with a score of 0.99, whereas the splice score for the mutated sequence c.1584+4A>T falls to 0.44. We hypothesized that the mutation leads to the activation of a cryptic splice site, with a score of 0.89, 63 bp downstream of the original splice site (Figure 2a). Indeed, RT-PCR confirmed that the mutated splice donor site induces the formation of a splice variant with 63 additional nucleotides from intron 7. This leads to an in-frame insertion of 21 amino acids (p.E528_V529ins21) after the amino acids encoded by exon 7. The wild-type sequence is correctly spliced in the control individual (Figure 2b).
Identification of the c.1584+4A>T disease-causing splice-site mutation in LRP5. (a) Direct-sequencing results of genomic DNA amplicons of the LRP5 exon 7 splice donor site in a control individual (wild-type sequence) and patient 3A (mutated sequence). The predicted splice score (0.99) of the wild-type splice donor site decreases to 0.44 in the mutated sequence, inducing the activation of a cryptic splice donor site downstream. (b) Electropherograms of LRP5 cDNA amplicons from patient 3A (upper panel) and a control individual (lower panel) revealing a mutated splice donor site after exon 7, with 63 additional nucleotides of intron 7 inserted into the RNA molecule in the patient's sample. The wild-type sequence is correctly spliced in the control individual.
LRP5 serves as a coreceptor with members of the Frizzled protein family for transducing signals by Norrin and Wnt proteins in the canonical Wnt-pathway. We used the dual-luciferase reporter assay with a Firefly luciferase reporter gene construct (Topflash TCF reporter plasmid) to measure the activity of the canonical Wnt-pathway upon Wnt/Norrin binding to LRP5. The assay revealed severely impaired signal transduction of the mutated LRP5 receptor (Figure 3a). Western-blot experiments with whole-cell lysates demonstrated that the mutant protein is stable when overexpressed (Figure 3b). Proteins with a signal peptide and a transmembrane domain are cotranslationally inserted into the membrane of the endoplasmatic reticulum. To determine whether the mutated LRP5 receptor is correctly inserted into the membrane, we separated the membrane fraction from the cytosolic fraction of transiently transfected cells. Wild-type LRP5 and LRP5 with the 21-amino-acid insertion were both almost exclusively detectable in the membrane fraction. However, the mutant LRP5 was present in lower amounts (Figure 3c). As the western blots from whole-cell lysates (Figure 3b) provided no evidence for an ineffective production of the mutant LRP5, we interpreted this observation as an experimental artifact.
Functional studies with mutant LRP5 protein. (a) Dual-luciferase reporter assay measuring the canonical Wnt signaling activity. The co-transfected Firefly luciferase reportergen construct (Topflash TCF reporter plasmid) contains TCF-binding sites as a target for activated canonical Wnt signaling. The graph describes the relative luciferase activities (average and standard deviation) of empty vector (pcDNA3.1), wild-type LRP5 (LRP5-WT9L), and the mutant LRP5-p.E528_V529ins21. The mutant LRP5-Mut3L serves as a positive control for impaired Wnt/Norrin signal transduction.25 Note that activities of the mutant LRP5-p.E528_V529ins21 are significantly lower than those of LRP5-WT9 L (P-values <0.001 (***) in a two-sided Student's t-test calculation). (b) Western blot of cell lysates from transiently transfected HEK293T cells used in the dual-luciferase reporter assay (results presented in a), confirming expression of WT9L and of the two mutated LRP5 receptors. (c) Cell fractioning assay. HEK293T cells were transiently transfected with LRP5-WT9L or LRP5-p.E528_V529ins21. Both WT9L and p.E528_V529ins21 are almost exclusively detectable in the membrane fraction. (d) Secretion assay. Representative western blot of conditioned medium (CM) and cell lysate (LY) from HEK293T cells transiently transfected with LRP5N-WT9L, LRP5N-p.E528_V529ins21 and LRP5N-Mut3L expression constructs. Comparable loading of cell lysate or conditioned medium across experiments is indicated by immunodetection of β-actin or by Coomassie staining of a 60-kDa protein in the conditioned medium, respectively. Note that only LRP5N-WT9L is strongly detected in conditioned medium.
To further track the molecular basis of the LRP5 loss-of-function, we generated a construct that expressed LRP5 with the 21-amino-acid insertion, but without the receptor's transmembrane and cytosolic domains. Subsequently, we performed a secretion assay (Figure 3d): LRP5 variants lacking the transmembrane domain (LRP5N) are secreted into the medium when normally trafficked within the cell and can be detected there by western blotting. The mutant LRP5N-p.E528_V529ins21 was almost completely absent from the medium, indicating that the intracellular transport of the receptor is disturbed.
Sequencing of genomic DNA in patient 4 confirmed homozygosity for common LRP5 SNPs, without identification of any putative disease-causing mutations. The PCR amplification of exon 4 with several different primer combinations was repeatedly unsuccessful in this patient, but always possible in DNA probes serving as controls. The sequencing of cDNA revealed the loss of exons 4 and 5 (Figure 4a). As we were able to amplify exons 3, 5 and part of intron 3 from genomic DNA of this patient (Figure 4b), a homozygous deletion involving exon 4 and part of the adjacent introns is likely. The amplification of a PCR fragment spanning the breakpoints has been unsuccessful.
(a) Electropherograms of LRP5 cDNA amplicons from patient 4 revealed the loss of exons 4 and 5. (b) PCR-amplified fragments from genomic DNA of patient 4 and two controls visualized on a 2% ethidium bromide gel. All studied amplicons; exons 3, 4 and 5, areas of intron 3 (a: 2735–3671 nucleotides and b: 4198–4503 nucleotides downstream of exon 3) and intron 4 (607–1213 nucleotides downstream of exon 4) can be detected in both control samples (C) and are clean in water samples (0) serving as negative controls. PCR of the patient's (P) genomic DNA was repeatedly unsuccessful in the areas of intron 3b, exon 4 and intron 4, suggesting a homozygous deletion involving exon 4 and adjacent intronic parts. Exon 5 and at least 10 nucleotides of the adjoining introns could be sequenced from both patient and control samples.
Discussion
OPPG is a rare autosomal recessive syndrome; only ∼60 patients have been reported worldwide. The phenotype in our patients is typical for OPPG, with loss of vision in infancy and severe symptomatic osteoporosis presenting in childhood as vertebral compression fractures and long bone fractures from minimal trauma. According to previous reports, heterozygous mutation carriers also exhibit a bone phenotype.6, 7, 27, 28, 29 In addition, recent studies on LRP5 polymorphisms and genome-wide association studies have revealed that common sequence variants in LRP5 influence BMD in the general population.30, 31, 32, 33, 34, 35 The majority of mutations linked to OPPG are found in the second and third of the four YTWD β-propeller domains, coded for by exons 6 to 12. Nearly half of the hitherto-published loss-of-function LRP5 mutations are missense mutations, one-fourth are nonsense mutations and one-fourth are frameshift mutations. Only 5% result in abnormal splicing. Of the known splice mutations, three have been associated with OPPG and one with FEVR; the bone phenotype in the FEVR patient is unknown.1 To our knowledge, the effect of human LRP5 splice aberrations on the function of the respective LRP5 protein have not been assessed experimentally so far. In this paper, we expand the spectrum of disease-causing mutations in OPPG by reporting novel splice mutations and the resulting phenotypes.
Two of the mutations affect the immediate and highly conserved splice site (±2 bases from exons) and one activates a cryptic splice site. The fourth mutation is a deletion causing the loss of two exons. The exact breakpoints have not been identified. The lack of tissue samples and RNA from patients 1 and 2 hindered the use of RT-PCR and expression studies, however, the implications of the mutations were visualized using a splicing assay. Both the substitution of the splice acceptor site (AG to GG) and of the donor site (GT to TT) are common, well-documented causes of exon skipping.36 The heterologous splicing assay confirmed exon skipping in our patients and excluded the formal possibility that cryptic splice sites could be used. The in-frame splicing-out of exon 20 in patient 1 does not form a stop codon but completely removes the transmembrane domain of the protein and hinders its function as a cell-surface signaling receptor (Figure 5). The mutant protein encoded from the allele harboring the S356 L mutation, although able to traffic normally, is unable to transduce Wnt1 and Wnt10b signals.1 The in-frame splicing-out of exon 5 in patient 2 introduces a premature stop codon, generated by the junction of exon 4 and exon 6, corresponding to amino-acid position 295. The truncation of both alleles at the start of exon 5 could explain the patient’s severe phenotype and mental retardation. However, normal cognitive development has been reported in other homozygous OPPG patients with more severe protein truncation, implying that additional factors influence mental development.1 No specific domain with regard to genotype–phenotype correlation and mental retardation can be found; however, the majority of mutations linked to mental retardation induce truncation of the protein. Retardation has been reported in both homozygous and compound heterozygous OPPG patients.1
Schematic representation of the LRP5 protein and the areas affected by the mutations presented in this study. (1) The splicing-out of exon 20 in patient 1 abolishes the entire transmembrane domain of the protein. (2) The in-frame splicing-out of exon 5 in patient 2 induces a stop codon at the beginning of exon 5, which encodes the first EGF repeat. (3) The insertion of 21 amino acids after exon 7 in patient 3 affects the second β-propeller between two YTWD domains. (4) The loss of exons 4 and 5 in patient 4 causes the loss of the last YWTD domain in the first β-propeller and the entire first EGF repeat.
The splice-site mutation c.1584+4A>T in patient 3 resulted in the insertion of 21 amino acids into the second β-propeller of the extracellular part of LRP5 (Figure 5). The same insertion has recently been found in a Japanese patient who carries a different mutation affecting the same splice site: c.1584+1G>A.19 Both these mutations inactivate the donor splice site, which activates the same cryptic splice site downstream. Our experimental data obtained, among others, with artificially truncated forms of LRP5 lacking the transmembrane and cytosolic domains, suggest that the insertion causes a disruption of intracellular protein trafficking. This interpretation is in-line with the severely reduced signal transduction we observed for the mutant LRP5 receptor in a luciferase reporter assay. A possible explanation for impaired trafficking is a conformational change in the receptor interfering with molecular interactions required for proper transport to the cytoplasmic membrane. Two other LRP5 mutations that interfere with protein trafficking have been described. One abolished the function of LRP5 signal peptide and hence prevented the transport of the nascent polypeptide chain to the endoplasmatic reticulum.25 This mechanism could be excluded for the 21 amino-acid insertion reported here. The molecular mechanism responsible for defective trafficking for the other reported LRP5 mutation (T390K) is unknown.1 However, the mutation in patient 3 is to our knowledge the first splice mutation in LRP5 of which the cellular consequences have been described with functional studies. Splice-site mutations at position +4 are rare, accounting for less than 2% of all donor-site mutations.37
In patient 4, we were not able to pinpoint the disease-causing sequence change on DNA level, but the evidence suggests a genomic deletion involving exon 4 and parts of introns 3 and 4. The sequencing of cDNA of the patient revealed loss of exons 4 and 5, which is likely to significantly impair the protein function and confirms the involvement of LRP5 in the patient's disorder.
All six heterozygous carriers who had undergone bone densitometry in our study had low BMD; one was diagnosed with osteoporosis and five with osteopenia. This is in accordance with previously published data showing heterozygous LRP5 mutations to have a dominant negative effect on BMD.12 One parent has adult-onset insulin-dependent diabetes mellitus. The link between LRP5 mutations, hyperglycemia through β-cell dysfunction and dyslipidemia has been investigated in animal studies and in a large OPPG family.12, 38 Diabetes does not seem to be a common complication in OPPG; however, glucose tolerance testing has not been performed in most patients or their parents. Our previous study showed diabetes in 50% (6 of 12) and abnormal glucose tolerance in 58% (7 of 12) of those harboring LRP5 mutation(s). The corresponding numbers in mutation-negative family members were 17% (one of six) and 33% (two of six).
Intronic changes in LRP5 may be overlooked with DNA screening, and deletions can be difficult to diagnose properly without RNA, MLPA or oligonucleotide tiling microarray studies.19 This is illustrated by patient 4 in whom we have not yet been able to characterize the exact boundaries of the homozygous genomic deletion. The novel mutations reported here expand the spectrum of genetic pathology underlying OPPG. The molecular mechanisms resulting in the variable phenotypic presentation remain to be elucidated in future studies.
References
Ai M, Heeger S, Bartels CF, Schelling DK : Clinical and molecular findings in osteoporosis-pseudoglioma syndrome. Am J Hum Genet 2005; 77: 741–753.
Gong Y, Vikkula M, Boon L et al: Osteoporosis-pseudoglioma syndrome, a disorder affecting skeletal strength and vision, is assigned to chromosome region 11q12–13. Am J Hum Genet 1996; 59: 146–151.
Somer H, Palotie A, Somer M, Hoikka V, Peltonen L : Osteoporosis-pseudoglioma syndrome: clinical, morphological, and biochemical studies. J Med Genet 1988; 25: 543–549.
Beighton P : Osteoporosis-pseudoglioma syndrome. Clin Genet 1986; 29: 263.
Frontali M, Dallapiccola B : Osteoporosis-pseudoglioma syndrome and the ocular form of osteogenesis imperfecta. Clin Genet 1986; 29: 262.
Lev D, Binson I, Foldes AJ, Watemberg N, Lerman-Sagie T : Decreased bone density in carriers and patients of an Israeli family with the osteoporosis-pseudoglioma syndrome. Isr Med Assoc J 2003; 5: 419–421.
Gong Y, Slee RB, Fukai N et al: LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell 2001; 107: 513–523.
Barros ER, Dias da Silva MR, Kunii IS, Hauache OM, Lazaretti-Castro M : A novel mutation in the LRP5 gene is associated with osteoporosis-pseudoglioma syndrome. Osteoporos Int 2007; 18: 1017–1018.
Cheung WM, Jin LY, Smith DK et al: A family with osteoporosis pseudoglioma syndrome due to compound heterozygosity of two novel mutations in the LRP5 gene. Bone 2006; 39: 470–476.
Crabbe P, Balemans W, Willaert A et al: Missense mutations in LRP5 are not a common cause of idiopathic osteoporosis in adult men. J Bone Miner Res 2005; 20: 1951–1959.
Streeten EA, McBride D, Puffenberger E et al: Osteoporosis-pseudoglioma syndrome: description of 9 new cases and beneficial response to bisphosphonates. Bone 2008; 43: 584–590.
Saarinen A, Saukkonen T, Kivela T et al: LDL receptor-related protein 5 (LRP5) mutations and osteoporosis, impaired glucose metabolism and hypercholesterolaemia. Clin Endocrinol (Oxf) 2010; 72: 481–488.
Toomes C, Bottomley HM, Jackson RM et al: Mutations in LRP5 or FZD4 underlie the common familial exudative vitreoretinopathy locus on chromosome 11q. Am J Hum Genet 2004; 74: 721–730.
Van Wesenbeeck L, Cleiren E, Gram J et al: Six novel missense mutations in the LDL receptor-related protein 5 (LRP5) gene in different conditions with an increased bone density. Am J Hum Genet 2003; 72: 763–771.
Boyden LM, Mao J, Belsky J et al: High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med 2002; 346: 1513–1521.
Little RD, Carulli JP, Del Mastro RG et al: A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am J Hum Genet 2002; 70: 11–19.
Westendorf JJ, Kahler RA, Schroeder TM : Wnt signaling in osteoblasts and bone diseases. Gene 2004; 341: 19–39.
Yadav VK, Ryu JH, Suda N et al: Lrp5 controls bone formation by inhibiting serotonin synthesis in the duodenum. Cell 2008; 135: 825–837.
Narumi S, Numakura C, Shiihara T et al: Various types of LRP5 mutations in four patients with osteoporosis-pseudoglioma syndrome: identification of a 7.2-kb microdeletion using oligonucleotide tiling microarray. Am J Med Genet A 2010; 152A: 133–140.
Wenstrup RJ, Florer JB, Willing MC et al: COL5A1 haploinsufficiency is a common molecular mechanism underlying the classical form of EDS. Am J Hum Genet 2000; 66: 1766–1776.
Buckler AJ, Chang DD, Graw SL et al: Exon amplification: a strategy to isolate mammalian genes based on RNA splicing. Proc Natl Acad Sci USA 1991; 88: 4005–4009.
Korinek V, Barker N, Morin PJ et al: Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science 1997; 275: 1784–1787.
Xu Q, Wang Y, Dabdoub A et al: Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell 2004; 116: 883–895.
Hsieh JC, Lee L, Zhang L et al: Mesd encodes an LRP5/6 chaperone essential for specification of mouse embryonic polarity. Cell 2003; 112: 355–367.
Chung BD, Kayserili H, Ai M et al: A mutation in the signal sequence of LRP5 in a family with an osteoporosis-pseudoglioma syndrome (OPPG)-like phenotype indicates a novel disease mechanism for trinucleotide repeats. Hum Mutat 2009; 30: 641–648.
Marsh M, Schmid S, Kern H et al: Rapid analytical and preparative isolation of functional endosomes by free flow electrophoresis. J Cell Biol 1987; 104: 875–886.
Downey LM, Bottomley HM, Sheridan E et al: Reduced bone mineral density and hyaloid vasculature remnants in a consanguineous recessive FEVR family with a mutation in LRP5. Br J Ophthalmol 2006; 90: 1163–1167.
Hartikka H, Makitie O, Mannikko M et al: Heterozygous mutations in the LDL receptor-related protein 5 (LRP5) gene are associated with primary osteoporosis in children. J Bone Miner Res 2005; 20: 783–789.
Bayram F, Tanriverdi F, Kurtoglu S et al: Effects of 3 years of intravenous pamidronate treatment on bone markers and bone mineral density in a patient with osteoporosis-pseudoglioma syndrome (OPPG). J Pediatr Endocrinol Metab 2006; 19: 275–279.
Baldock PA, Eisman JA : Genetic determinants of bone mass. Curr Opin Rheumatol 2004; 16: 450–456.
Ferrari SL, Deutsch S, Choudhury U et al: Polymorphisms in the low-density lipoprotein receptor-related protein 5 (LRP5) gene are associated with variation in vertebral bone mass, vertebral bone size, and stature in whites. Am J Hum Genet 2004; 74: 866–875.
Koay MA, Tobias JH, Leary SD, Steer CD, Vilarino-Guell C, Brown MA : The effect of LRP5 polymorphisms on bone mineral density is apparent in childhood. Calcif Tissue Int 2007; 81: 1–9.
Koay MA, Woon PY, Zhang Y et al: Influence of LRP5 polymorphisms on normal variation in BMD. J Bone Miner Res 2004; 19: 1619–1627.
Saarinen A, Valimaki VV, Valimaki MJ et al: The A1330V polymorphism of the low-density lipoprotein receptor-related protein 5 gene (LRP5) associates with low peak bone mass in young healthy men. Bone 2007; 40: 1006–1012.
Sims AM, Shephard N, Carter K et al: Genetic analyses in a sample of individuals with high or low BMD shows association with multiple Wnt pathway genes. J Bone Miner Res 2008; 23: 499–506.
Baralle M, Baralle FE : Genetics and molecular biology: variations in alternative spliced pre-mRNA-protein isoforms and their role in disease. Curr Opin Lipidol 2008; 19: 429–430.
Krawczak M, Thomas NS, Hundrieser B et al: Single base-pair substitutions in exon-intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum Mutat 2007; 28: 150–158.
Fujino T, Asaba H, Kang MJ et al: Low-density lipoprotein receptor-related protein 5 (LRP5) is essential for normal cholesterol metabolism and glucose-induced insulin secretion. Proc Natl Acad Sci USA 2003; 100: 229–234.
Acknowledgements
We thank our patients and their families for participating in this study and Bernadette C Holdener as well as Matthew L Warman for sharing reagents. We are grateful to Dr Dieter Meschede, Cologne, Germany, for referring patients 3A and 3B to us. This study was supported by Folkhälsan Institute of Genetics, Academy of Finland, Sigrid Juselius Foundation and Helsinki University Hospital Research Funds, and by a grant of the Deutsche Forschungsgemeinschaft (DFG) to CN (grant NE 826/3-2).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Laine, C., Chung, B., Susic, M. et al. Novel mutations affecting LRP5 splicing in patients with osteoporosis-pseudoglioma syndrome (OPPG). Eur J Hum Genet 19, 875–881 (2011). https://doi.org/10.1038/ejhg.2011.42
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2011.42
Keywords
This article is cited by
-
The implications of alternative pre-mRNA splicing in cell signal transduction
Experimental & Molecular Medicine (2023)
-
Osteoporosis-pseudoglioma syndrome in four new patients: identification of two novel LRP5 variants and insights on patients’ management using bisphosphonates therapy
Osteoporosis International (2022)
-
Competitive blocking of LRP4–sclerostin binding interface strongly promotes bone anabolic functions
Cellular and Molecular Life Sciences (2022)
-
Citrullinemia type I is associated with a novel splicing variant, c.773 + 4A > C, in ASS1: a case report and literature review
BMC Medical Genetics (2019)
-
Osteoporosis-pseudoglioma syndrome: clinical, genetic, and treatment-response study of 10 new cases in Greece
European Journal of Pediatrics (2019)