Abstract
Tuberous sclerosis complex (TSC) is a genetic condition characterized by the growth of benign tumours in multiple organs, including the brain and kidneys, alongside intellectual disability and seizures. Identification of a causative mutation in TSC1 or TSC2 is important for accurate genetic counselling in affected families, but it is not always clear from genetic data whether a sequence variant is pathogenic or not. In vitro functional analysis could provide support for determining whether an unclassified TSC1 or TSC2 variant is disease-causing. We have performed a detailed functional analysis of four patient-derived TSC2 mutations, E92V, R505Q, H597R and L1624P. One mutant, E92V, functioned similarly to wild-type TSC2, whereas H597R and L1624P had abnormal function in all assays, consistent with available clinical and segregation information. One TSC2 mutation, R505Q, was identified in a patient with intellectual disability, seizures and autistic spectrum disorder but who did not fulfil the diagnostic criteria for TSC. The R505Q mutation was also found in two relatives, one with mild learning difficulties and one without apparent phenotypic abnormality. R505Q TSC2 exhibited partially disrupted function in our assays. These data highlight the difficulties of assessing pathogenicity of a mutation and suggest that multiple lines of evidence, both genetic and functional, are required to assess the pathogenicity of some mutations.
Similar content being viewed by others
Introduction
Tuberous sclerosis complex (TSC) is an autosomal dominant condition characterized by the development of benign tumours in multiple organs, including the brain, skin, heart, lungs and kidneys, as well as neuropsychological manifestations including seizures, intellectual disability and autistic spectrum disorder. Mutations affecting TSC1 (on chromosome 9q34)1 and TSC2 (on chromosome 16p13.3)2 account for at least 80% of all TSC cases.3 TSC2 mutations tend to result in a more severe phenotype.3, 4, 5 One of the factors contributing to the severity of the disease may be the position of the mutation and its functional effects on the protein product.
Some 1700 or so different variants affecting TSC1 and TSC2 have been reported.6 C-terminal truncations due to frameshift mutations and premature stop codons are observed commonly within TSC27 and missense mutations are distributed throughout the TSC2 coding sequence, accounting for the pathogenic change in ∼25 % of patients.7 By comparison, the vast majority of pathogenic TSC1 mutations appear to be truncating lesions and only a small number of TSC-causing missense mutations are reported.3 It is often difficult to predict the effects of missense mutations on protein function. Investigators have used several approaches to establish the pathogenicity of TSC missense variants (Supplementary Table 1). There are problems with all of these approaches. Parental samples may not be available or family structure may not be suitable to draw any definite conclusions from segregation analysis. Protein alignments can have too little or too much variation to arrive at useful conclusions from in silico analysis. The Practice Guidelines8 of the UK Clinical Molecular Genetics Society recommend that conclusions drawn from clinical genetic or in silico approaches should be based on more than one line of evidence. A reliable functional assay could greatly facilitate the interpretation of variants of uncertain clinical significance. Few TSC1 or TSC2 mutations have been characterized functionally9, 10, 11 and it is still impossible to predict phenotypic effects from the location and type of mutation.
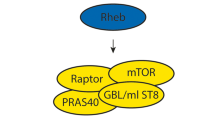
TSC1 (hamartin) and TSC2 (tuberin) proteins are considered to act as tumour suppressors. As a heterodimer, TSC1 and TSC2 repress cell growth by inhibiting cell signal transduction through the mammalian target of rapamycin complex 1 (mTORC1) pathway.12 TSC1/TSC2 possesses GTPase-activating protein (GAP) activity towards the small G-protein, Ras homologue enriched in brain (Rheb),13, 14, 15, 16 converting Rheb from its active GTP-bound form to the inactive GDP-bound form. Consequently, inactivation of Rheb by TSC1/TSC2 prevents activity of mTORC1 and phosphorylation of the mTORC1 substrates 4E-BP1 and S6K1, leading to repression of protein translation. Mutations within either TSC1 or TSC2 disrupt the function of the heterodimer, allowing mTORC1 to be inappropriately active, increasing cell growth and proliferation.
We have analysed the consequences of missense mutations in four highly conserved regions of TSC2 (of both known and unknown function), recreated from mutations observed in patients referred for diagnostic molecular genetic testing of TSC1 and TSC2. Three remain unique mutations, while the other, E92V, has now been detected in two patients. We have analysed the stability of these TSC2 mutants, as well as their ability to function as a GAP towards Rheb and their capacity to repress mTORC1 signalling via analysis of the mTORC1 targets, 4E-BP1, S6K1 and hypoxia inducible factor 1α (HIF-1α).
Materials and methods
Patients and mutation screening
TSC2 missense variants were selected for analysis from a database of mutations identified in patient samples referred to the clinical genetics service laboratory in Medical Genetics (Cardiff University) for TSC1 and TSC2 mutation analysis because of a possible diagnosis of TSC. TSC1 and TSC2 mutation screening was performed using denaturing high-performance liquid chromatography (DHPLC), essentially as described in Jones et al.17 Acetylnitrile gradients were modified for the WAVE 3500HT instrumentation and HT column to reduce the run time to 3 min per sample (contact authors for details). The TSC PCR products were sequenced as described below. Exonuclease I (New England Biolabs, Hitchen, Hertfordshire, UK) and shrimp alkaline phosphatase (Amersham Biosciences, Little Chalfont, Buckinghamshire, UK) were combined in equal volumes and 1 μl of this mix was added to 3 μl of PCR product. The reactions were performed in a thermal cycler (9700, Applied Biosystems, Warrington, UK) using the following cycle: 37°C for 15 min, 80°C for 15 min, hold at 4°C. A mastermix of 2 μl of 5 × sequencing buffer, 0.75 μl BigDye Terminator v3.1 Sequencing Kit (Applied Biosystems) and 5.25 μl of water per reaction was prepared. A volume of 8 μl of mastermix was combined with 1 μl of exo/SAP-treated PCR product and 1 μl of the appropriate sequencing primer diluted to 1.6 pmol/μl. The primers used covered all of the coding exons of both TSC1 and TSC2. The following PCR cycle was used: 96°C for 10 s, 50°C for 5 s and 60°C for 4 min for 25 cycles. Subsequently, BigDye terminators were removed using the Montage SEQ96 sequencing reaction cleanup kit (Millipore, Co., Durham, UK) as described in the manufacturer's instructions. The sequencing reactions were run on an ABI 3100 Genetic Analyser. Multiplex ligation-dependent probe amplification (MLPA) was performed on TSC2 using a standard kit, P046-B2 (MRC Holland, Amsterdam, The Netherlands). A questionnaire documenting symptoms, signs and radiographic findings was completed for each patient by the referring clinician.
Plasmid details
Flag-tagged TSC2 wild-type vector was a kind gift of Dr C Walker (MD Anderson Center, Houston, TX, USA). Site-directed mutagenesis of the wild-type vector was used to create the mutants E92V, R505Q, H597R and L1624P. All site-directed mutagenesis was performed using Phusion DNA polymerase (New England Biolabs (UK) Ltd) and TOP 10 cells (Invitrogen, Paisley, UK). TSC1 was amplified by PCR from a pRK7/Flag-TSC1 vector and subcloned into the pcDNA3.1/n-V5-DEST vector using the Gateway system (Invitrogen). GST-Flag-Rheb and HA-S6K1 were kindly provided by Prof J Blenis (Harvard Medical School, Boston, MA, USA). C-terminally tagged Myc-4E-BP1 was generated by digesting PCR fragments encoding rat 4E-BP1 with HindIII and BamHI followed by cloning into pcDNA3.1myc/3his− (Invitrogen). The TSC2 wild-type and mutant constructs were subcloned into a GFP-tagged vector using the Gateway system. TSC2 vectors were sequence verified (MWG Biotech, Ebersberg, Germany).
Antibodies and other biochemicals
The antibodies used to perform western blotting were clone 9B11 anti-Myc (Roche, Welwyn Garden City, UK), anti-phospho S6 Kinase 1 (Thr389), total S6 Kinase 1, anti-TSC2 and anti-phospho-4E-BP1 Ser65 (Cell Signaling, Hitchen, Hertfordshire, UK), anti-Rheb (Santa Cruz, Heidelberg, Germany), anti-Flag M2 (Sigma, Dorset, UK) and anti-V5 (Invitrogen). Insulin and cycloheximide were purchased from Sigma.
Cell culture and transfection
HEK293E cells and Tsc2−/− mouse embryonic fibroblasts (MEFs) were cultured in Dulbecco's modified Eagle's medium supplemented with 10 % fetal calf serum, 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco, Paisley, UK). CaCl2-mediated transfection was carried out as described previously.18 Lipofectamine 2000 (Invitrogen) transfections were performed according to the manufacturer's protocol. Cells were either co-transfected with Flag-tagged TSC2 (wild type or mutant) and Flag-tagged or V5-tagged TSC1 constructs with or without a GST-tagged Rheb construct. For 4E-BP1 experiments, Myc-tagged 4E-BP1 was also co-transfected, whereas for S6K1 assays, HA-S6K1 was co-transfected. At 24 h post-transfection, after overnight serum-starvation, these cells were harvested (10 mM potassium phosphate, 1 mM EDTA, 5 mM EGTA, 10 mM MgCl2, 50 mM β-glycerophosphate, 0.5% (vol/vol) NP40, 0.1% (vol/vol) Brij-35, pH 7.4, plus protease inhibitors). Cells requiring insulin stimulation were treated with 10 μg/ml insulin for 30 min before lysis. Cells requiring cycloheximide treatment were treated with 10 μg/ml cycloheximide for the indicated time points before harvesting. Cell lysates for subsequent immunoprecipitation were lysed in a different buffer, as detailed below.
Immunoprecipitations
Cells were lysed in an NP40-based buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 1 mM MgCl2, 50 mM β-glycerophosphate, 10% (vol/vol) glycerol, 1% (vol/vol) NP40 plus protease inhibitors) and centrifuged at 13 000 r.p.m. for 8 min at 4°C to remove insoluble matter. The supernatant was precipitated using anti-Flag M2 antibody and Protein G-sepharose beads (GE Healthcare, Little Chalfont, Buckinghamshire, UK). Beads were washed twice in lysis buffer, then twice in wash buffer (20 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM EDTA, 50 mM NaF, 50 mM β-glycerophosphate, 1% (vol/vol) NP40) before resuspension in sample buffer (Invitrogen).
Western blotting
Cell lysates were resolved by SDS-PAGE and proteins were transferred to polyvinylidene difluoride membranes (Millipore). Membranes were blocked in 5% (wt/vol) dry milk powder dissolved in Tris-buffered saline containing 0.1% (vol/vol) Tween, then probed using the required primary antibody and horse radish peroxidase-conjugated secondary antibody (Sigma). Proteins were visualized using Enhanced Chemiluminescent solution and Hyperfilm (both GE Healthcare). All western blots shown are representative of at least three independent experiments. Densitometry was performed using ImageJ software (National Institutes of Health, http://rsbweb.nih.gov/ij/index.html) and the resulting data analysed using Microsoft Excel.
S6K1 assay and HIF1α activity assay
These assays were performed as described previously,19 using recombinant GST-rpS6 (32 C-terminal amino acids of ribosomal protein S6) as substrate or using the firefly luciferase reporter pGL2-TK-HRE plasmid to report HIF activity. This construct was generated by subcloning three copies of the hypoxia response element (HRE; 5′-GTGACTACGTGCTGCCTAG-3′) from the inducible nitric oxide synthase promoter into the promoter region of the pGL2-TK vector and was kindly provided by G Melillo (National Cancer Institute, Frederick, MD, USA).
GAP assay
Immunocomplexes of Flag-tagged TSC1 and TSC2 were subjected to in vitro RhebGAP assays as previously described.16 α-[32P]GTP and α-[32P]GDP were eluted from Rheb and resolved by thin layer chromatography on PEI cellulose (Sigma) with 1 M KH2PO4, pH 3.4. The relative levels of [32P]-radiolabelled GTP and GDP were determined by autoradiography.
GFP-TSC2 localization
GFP-tagged TSC2 constructs were transfected into Tsc2−/− MEFs using Lipofectamine 2000 (Invitrogen) and then serum-starved for 16 h. The distribution of GFP fluorescence was detected using a Zeiss LSM510 confocal microscope (Zeiss, Welwyn Garden City, UK). Note that identical microscope settings were employed for each TSC2 construct analysed.
Results
In silico analysis of TSC2 mutants
We investigated four previously uncharacterized patient-derived mutations of TSC2, spread across the length of the TSC2 gene: E92V, R505Q, H597R and L1624P (Figure 1a). These were the only significant changes in the patients investigated. The DHPLC diagnostic mutation screen used was previously validated using a patient panel of formerly identified mutations. Where a known point mutation was present, 97% of mutations were identified by DHPLC.3 No deletions in TSC2 were detected using MLPA. All of the mutations are located in highly conserved regions of the TSC2 protein, although not all are in domains with known functions. The mutated residues are highly conserved between species (Supplementary Table 2). In silico analysis using the computer prediction programmes Polyphen (http://genetics.bwh.harvard.edu/pph/) and SIFT (http://blocks.fhcrc.org/sift/SIFT.html) suggested that R505Q was unlikely to significantly affect protein structure/function but that the other mutations might (Table 1, upper panel). Using Alamut (www.interactivebiosoftware.com/alamut.html), the effect of the mutations on splicing was investigated. No significant effect on splicing was predicted for any of the mutations.
(a) A schematic of the TSC2 protein showing the key functional domains and major phosphorylation sites. The patient-derived mutations are shown above the protein structure. (b) Stability of the TSC2 mutant proteins transiently transfected into HEK293 cells was analysed following 10 μg/mg cycloheximide treatment for 9 h. Total cell lysates were probed with anti-Flag antibody. The western blots shown are representative of three independent experiments, while the densitometry data for the level of TSC2 protein shows the mean and standard deviation of all three experiments. The 0- and 9-h time points were compared using a t-test, and P-values are indicated: NS, not significant; *P<0.05; **P<0.01.
Clinical features of patients
The phenotypic details of the patients are given in Table 2. Patients 1, 3 and 4 fulfilled diagnostic criteria for TSC.20 The missense mutation carried by patient 1 (E92V) was also present in her apparently unaffected mother. Patient 2 carried the R505Q mutation and had intellectual disability, autistic spectrum features, seizures and facial lesions suggestive of angiofibroma (not biopsied). Although investigations for all possible manifestations of TSC had not been undertaken, this patient did not fulfil diagnostic criteria for TSC. The mutation was also found in her mother, who was reported to have mild intellectual disability but no other clinical manifestations of TSC, and her apparently unaffected grandmother, neither of whom were fully assessed for features of TSC. The H597R mutation carried by patient 3 was not present in his clinically unaffected mother, but no DNA sample was available from his father for testing. The L1624P mutation found in patient 4 was not present in either of his clinically phenotypically unaffected parents.
Stability of the TSC1/TSC2 complex in the presence of TSC2 missense mutations
To measure protein stability, we analysed loss of wild-type and mutant TSC2 protein over a 9-h time course after blocking de novo protein synthesis with cycloheximide. Both wild-type and E92V TSC2 remained fairly stable over the 9-h cycloheximide time course. However, there was significant loss of the other three mutant proteins after 9 h of cycloheximide treatment. The level of TSC2 protein within cell lysates was reduced by 56±13% with the R505Q mutant, 79±2% with the H597R mutant and 45±13% with the L1624P mutant (Figure 1b), as shown by the densitometry data. Although TSC1 protein levels did not significantly alter over the course of the treatment, the total TSC1 protein level detectable when co-expressed with the R505Q and H597R mutants was reduced (Figure 1b, central panels of TSC1 blots).
TSC1/TSC2 heterodimer formation and in vitro RhebGAP activity
The cell signalling network we analysed to further characterize the protein function of these TSC2 mutants is depicted in Figure 2a. Analysis included: (i) interaction of TSC2 with TSC1; (ii) in vitro RhebGAP activity, and an examination of signal transduction through mTORC1 to three downstream substrates; (iii) S6K1, (iv) 4E-BP1 and (v) HIF-1α. As TSC2 forms a functional heterodimer with TSC1, we wanted to determine whether the patient-derived TSC2 mutations affected interaction with TSC1. Co-immunoprecipitation studies were performed on TSC2 and the level of associated TSC1 was determined (Figure 2b). TSC2 E92V interacted with TSC1 at a comparable level to wild-type TSC2 protein. The other three TSC2 mutants (R505Q, H597R and L1624P) showed >50% reduction in binding to TSC1, perhaps due to conformational changes or because of increased targeting of TSC2 for degradation.
(a) A diagram of the cell signalling network analysed, which includes (i) the interaction of TSC2 with TSC1, (ii) an in vitro RhebGAP activity assay, and assays for (iii) S6K1, (iv) 4E-BP1 and (v) HIF-1α to measure downstream signalling through mTORC1. (b) Formation of the TSC1/TSC2 heterodimer was analysed by immunoprecipitation of Flag-TSC2 followed by western blotting for V5-TSC1. Immunoprecipitated Flag and total V5 blots are shown as controls. The percentage of TSC1 bound to TSC2 is shown relative to the wild-type binding level. All densitometry data were corrected for total TSC1 and TSC2 levels and are shown as the mean±SD of combined independent experiments. The interaction level of each mutant was compared with wild type using a t-test; *indicates P<0.01 (lower panel). (c) In vitro RhebGAP activity was assessed for each TSC2 mutant, and the levels of GTP and GDP following the assay were analysed by thin layer chromatography. The ratio of GTP/GDP for the representative experiment shown is below the autoradiograph. Combined densitometry data (mean±SD) for the conversion to GDP are shown in the graph below the blot, with those mutants showing a significant difference to wild-type indicated (*indicates P<0.01). Total TSC1 and TSC2 protein levels are shown as a loading control.
The GAP domain within TSC2 is responsible for activating the intrinsic GTPase activity of Rheb, thereby converting active GTP-bound Rheb into its inactive GDP-bound form. We assessed the in vitro RhebGAP activity of the TSC2 mutants and found that only wild-type TSC2 and the E92V mutant promoted the intrinsic GTPase activity of Rheb, as observed by hydrolysis of GTP to GDP. R505Q, H597R and L1624P had lost their ability to act as a GAP, although only L1624P is located within the GAP domain (Figure 2c).
Ability to inhibit phosphorylation of downstream substrates of mTORC1
We wanted to test whether loss of RhebGAP activity of the R505Q, H597R and L1624P mutants resulted in upregulation of the mTORC1 pathway. To do this we examined two mTORC1 substrates, S6K1 and 4E-BP1. We determined the activity of S6K1 and the phosphorylation state of 4E-BP1 when the TSC2 mutants were expressed. The E92V mutant behaved similarly to the wild type, inhibiting S6K1 activity under serum-starved conditions as indicated by the low level of phosphorylation of its substrate rpS6 (Figure 3a). Both wild-type and E92V TSC2 also repressed 4E-BP1 phosphorylation under serum-starved conditions that was reversed by insulin stimulation (Figure 3a). The H597R and L1624P mutants were unable to repress mTORC1 activity under serum-starved conditions to a level equivalent to wild type, as shown by the significantly increased S6K1 activity and the significantly increased phosphorylation of Ser65 in 4E-BP1. The increased level of phosphorylated 4E-BP1 associated with the H597R and L1624P mutants was also revealed by the presence of the hyperphosphorylated γ-species of 4E-BP1 (Figure 3a, bottom panel) and an increase in the proportion of the moderately phosphorylated β-isoform. The R505Q mutant did appear able to repress Rheb-induced S6K1 activity, as shown by low basal rpS6 phosphorylation but showed a significantly increased level of basal 4E-BP1 phosphorylation (Figure 3a).
(a) Activation of mTORC1 following TSC2 mutant overexpression was assessed by analysing downstream mTORC1 targets. An S6K1 assay was performed using a rpS6 peptide as substrate. Incorporation of [32P]-radiolabel into rpS6 was assessed by autoradiography, and S6K1 immunoprecipitates were probed for total S6K1. Blots shown are representative of three independent experiments (middle panels). HEK293 cells were transiently transfected with Flag-TSC1, Flag-TSC2 (wild type or mutant), Flag-Rheb and Myc-4E-BP1 and treated with or without 100 nM insulin for 30 min as shown in the figure. Total cell lysates were probed for phospho-Ser65 4E-BP1 and total 4E-BP1. Each isoform of 4E-BP1 in the total blot was assessed by densitometry and the percentage of each isoform is indicated under the figure. Blots shown are representative of three independent experiments (lower panel). Levels of TSC1, TSC2 and Rheb are shown as controls (upper panels). (b) Densitometry data for the levels of phospho-rpS6 (upper graph) and phospho-4E-BP1 at Ser65 (lower graph) are shown. Bars represent the mean±SEM of the combined data. The level of repression in unstimulated cells was compared with the control samples using Student's t-test, *P<0.05. (c) The ability of the mutant TSC2 proteins to downregulate HIF-1α activity was assessed using a HIF-1α reporter assay. The graph shows data from three independent experiments, each performed in triplicate, with error bars representing standard deviation. Values which are statistically different to wild-type are indicated, where *P<0.01. Total TSC2 protein levels are shown in the western blot, and the densitometry for the total TSC2 protein levels across the three experiments is shown in the graph on the right. Levels were not significantly different.
HIF-1α was recently identified as an mTORC1 substrate.21 We therefore assessed HIF-1α transcriptional activity following overexpression of the TSC2 mutants in Tsc2−/− MEFs in a TSC2 rescue experiment, using an HIF-1α reporter assay. We utilized dimethyloxalylglycin, an inhibitor of prolyl hydroxylase, to mimic hypoxia and found that addition of the mTORC1 inhibitor, rapamycin, reduced HIF-1α activity by >60%. In this rescue experiment, overexpression of wild-type TSC2 or the E92V mutant completely repressed the high transcriptional activity of HIF-1α. The R505Q mutant was as effective as rapamycin, whereas the H597R and L1624P mutants produced a 30–40% reduction in HIF-1α transcriptional activity (Figure 3b).
Finally, we analysed the localization of TSC2 mutants within the cell using GFP–TSC2 constructs. All TSC2 mutants were observed to form a distinct, punctuate pattern in the absence of serum, as previously observed,22 and we did not detect any substantial difference in localization of these TSC2 mutants when compared with wild type (Supplementary Figure 1).
The observed effects of the mutants in each assay are summarized in Table 1, lower panel.
Discussion
The TSC2 mutations examined showed a range of functional deficits. The E92V variant, located in a leucine-zipper region, has been reported in two patients (although clinical details on just one were available to us) and in silico analysis predicted that it may affect protein function. The apparently unaffected mother of patient 1 also carried the E92V variant suggesting that this mutation is unlikely to be pathogenic. The in vitro functional analyses reported here also indicated that pathogenicity is unlikely as the E92V protein behaved similarly to wild type. Therefore, we conclude that E92V is a neutral variant. E92V is the only variant that has been identified in this patient. No other changes were found using assays known to detect mutations in <90% of cases with a clinical diagnosis of TSC. It is possible that TSC in this patient could be due to deep intronic mutations affecting splicing or mutations in regulatory regions upstream of the start codon, as these regions will not have been screened.
Patient 2, who carried the R505Q mutation, had intellectual disability, autistic spectrum disorder, seizures and probable angiofibroma, but did not fulfil the diagnostic criteria for TSC on the basis of the investigations performed to date. The R505Q mutation was also found to be present in her mother, who was reported to have mild intellectual disability but no other clinical manifestations of TSC, and the mutation was also present in the proband's apparently normal grandmother. The computer models, Polyphen and SIFT, suggested this mutation would not affect protein function. However, our functional data suggest that this mutation confers at least partial loss of function on the protein. TSC2 R505Q was not found to have any in vitro GAP activity, nor could it repress Rheb-induced 4E-BP1 phosphorylation. Although it could repress HIF-1α activation to a similar extent to rapamycin, it was not as potent as wild-type TSC2. However, it did repress Rheb-induced S6K1 activity. The apparent disconnect between 4E-BP1 and S6K1 phosphorylation downstream of this mutant was surprising and may reflect different sensitivities of the assays used, or perhaps suggest subtle differences in control of phosphorylation of these two substrates by mTORC1 through Raptor interaction. Other groups have reported that Raptor binds much more weakly to S6K1 than 4E-BP123, 24 and we have made similar observations (data not shown). Unlike wild-type TSC2, it is possible that the R505Q mutant does not completely block mTORC1 signalling in cells. This could result in adequate 4E-BP1 phosphorylation through maintained Raptor association, while S6K1, which is a weaker Raptor binding target, may remain inactive. From the data available it remains unclear whether the R505Q mutation is truly pathogenic.
In silico analyses predicted that the H597R mutation identified in patient 3 with clinically confirmed TSC would affect protein function, and this was supported by the functional analyses, all of which indicated an inactive TSC2 protein. The limited segregation data available are compatible with the mutation being pathogenic. H597 does not lie within a known functional domain of TSC2, but is in a highly conserved region (Supplementary Figure 2). It remains to be determined how this region contributes to TSC2 function and how mutations within it can have such a deleterious effect on protein function.
The L1624P mutation identified in patient 4 with clinically confirmed TSC had arisen de novo, as it was not present in either parent. The mutation lies within the well-characterized GAP domain of TSC2, which has a direct action on the mTORC1 activator, Rheb. In silico analyses predicted it was likely to be harmful to protein function, and all functional analyses showed it to be defective in inhibiting mTORC1 signalling. Of interest, this mutant, which had no GAP activity and was unable to repress mTORC1, was still able to inhibit HIF by 32%. This highlights the possibility that TSC2 may inhibit the activity of HIF via both mTORC1-dependent and -independent mechanism(s).
The functional analyses of patient-derived TSC2 mutations described in this paper indicate that multiple assays may be required to fully characterize the functional consequences of some mutations. Recently, a semiautomated cell-based assay has been described to test the effects of unclassified TSC2 variants on protein function by looking at the ability of the mutants to repress S6K1 phosphorylation.11 Our findings of apparently separable effects of the R505Q mutation on S6K1 and 4E-BP1 phosphorylation suggest that caution may be required in the interpretation of single functional assays.
Functional analysis may help to assign pathogenicity to unclassified variants that are disease causing when other evidence is inconclusive.11 Confidence in interpreting results is increased when the conclusions from all lines of available evidence concur. However, the data obtained from functional analysis of the R505Q mutation highlight that functional data should be interpreted with caution until functional deficits and phenotypes have been better correlated in larger series. The Human Variome Project has the aim of collecting, curating and making accessible information on genetic variations affecting human health,25 and as researchers contribute their genetic and functional findings to databases such as LOVD, this should improve our knowledge of how different mutations affect protein function and cause disease.
References
European Chromosome 16 Tuberous Sclerosis Consortium: Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell 1993; 75: 1305–1315.
van Slegtenhorst M, de Hoogt R, Hermans C et al: Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science 1997; 277: 805–808.
Jones AC, Shyamsundar MM, Thomas MW et al: Comprehensive mutation analysis of TSC1 and TSC2-and phenotypic correlations in 150 families with tuberous sclerosis. Am J Hum Genet 1999; 64: 1305–1315.
Dabora SL, Jozwiak S, Franz DN et al: Mutational analysis in a cohort of 224 tuberous sclerosis patients indicates increased severity of TSC2, compared with TSC1, disease in multiple organs. Am J Hum Genet 2001; 68: 64–80.
Sancak O, Nellist M, Goedbloed M et al: Mutational analysis of the TSC1 and TSC2 genes in a diagnostic setting: genotype--phenotype correlations and comparison of diagnostic DNA techniques in Tuberous Sclerosis Complex. Eur J Hum Genet 2005; 13: 731–741.
Sue Povey and Rosemary Ekong (2005-2009). Tuberous sclerosis database. LOVD v.2.0, http://chromium.liacs.nl/LOVD2/TSC/home.php?select_db=TSC2.
Beauchamp RL, Banwell A, McNamara P et al: Exon scanning of the entire TSC2 gene for germline mutations in 40 unrelated patients with tuberous sclerosis. Hum Mutat 1998; 12: 408–416.
Bell J, Bodmer D, Sistermans E, Ramsden S : Practice guidelines for the Interpretation and Reporting of Unclassified Variants (UVs) in Clinical Molecular Genetics, 2007, www.cmgs.org/BPGs/pdfs%20current%20bpgs/UV%20GUIDELINES%20ratified.pdf.
Mozaffari M, Hoogeveen-Westerveld M, Kwiatkowski D et al: Identification of a region required for TSC1 stability by functional analysis of TSC1 missense mutations found in individuals with tuberous sclerosis complex. BMC Med Genet 2009; 10: 88.
Nellist M, van den Heuvel D, Schluep D et al: Missense mutations to the TSC1 gene cause tuberous sclerosis complex. Eur J Hum Genet 2009; 17: 319–328.
Coevoets R, Arican S, Hoogeveen-Westerveld M et al: A reliable cell-based assay for testing unclassified TSC2 gene variants. Eur J Hum Genet 2009; 17: 301–310.
Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, Blenis J : Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci USA 2002; 99: 13571–13576.
Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D : Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol 2003; 5: 578–581.
Garami A, Zwartkruis FJ, Nobukuni T et al: Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell 2003; 11: 1457–1466.
Inoki K, Li Y, Xu T, Guan KL : Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev 2003; 17: 1829–1834.
Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J : Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol 2003; 13: 1259–1268.
Jones AC, Sampson JR, Hoogendoorn B Cohen D, Cheadle JP : Application and evaluation of denaturing HPLC for molecular genetic analysis in tuberous sclerosis. Hum Genet 2000; 106: 663–668.
Schalm SS, Blenis J : Identification of a conserved motif required for mTOR signalling. Curr Biol 2002; 12: 632–639.
Dunlop EA, Dodd KM, Seymour LA, Tee AR : Mammalian target of rapamycin complex 1-mediated phosphorylation of eukaryotic initiation factor 4E-binding protein 1 requires multiple protein-protein interactions for substrate recognition. Cell Signal 2009; 21: 1073–1084.
Roach ES, Gomez MR, Northrup H : Tuberous Sclerosis Complex Consensus Conference: revised clinical diagnostic criteria. J Child Neurol 1998; 13: 624–628.
Land SC, Tee AR : Hypoxia-inducible factor 1alpha is regulated by the mammalian target of rapamycin (mTOR) via an mTOR signaling motif. J Biol Chem 2007; 282: 20534–20543.
Cai SL, Tee AR, Short JD et al: Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J Cell Biol 2006; 173: 279–289.
Schalm SS, Fingar DC, Sabatini DM, Blenis J : TOS Motif-mediated raptor binding regulates 4E-bp1multisite phosphorylation and function. Curr Biol 2003; 13: 797–806.
Hara K, Maruki Y, Long X et al: Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 2002; 110: 177–189.
Cotton RG, Auerbach AD, Axton M et al: The Human Variome Project. Science 2008; 322: 861–862.
Acknowledgements
This work was supported by the Association for International Cancer Research Career Development Fellowship (no. 06-914/915; to A Tee), the Tuberous Sclerosis Association and Wales Gene Park. We also thank the All Wales Medical Genetics Service.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Dunlop, E., Dodd, K., Land, S. et al. Determining the pathogenicity of patient-derived TSC2 mutations by functional characterization and clinical evidence. Eur J Hum Genet 19, 789–795 (2011). https://doi.org/10.1038/ejhg.2011.38
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2011.38