Abstract
Prader–Willi syndrome (PWS) is a multisystem, contiguous gene disorder caused by an absence of paternally expressed genes within the 15q11.2-q13 region via one of the three main genetic mechanisms: deletion of the paternally inherited 15q11.2-q13 region, maternal uniparental disomy and imprinting defect. The deletion class is typically subdivided into Type 1 and Type 2 based on their proximal breakpoints (BP1–BP3 and BP2–BP3, respectively). Despite PWS being a well-characterized genetic disorder the role of the specific genes contributing to various aspects of the phenotype are not well understood. Methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA) is a recently developed technique that detects copy number changes and aberrant DNA methylation. In this study, we initially applied MS-MLPA to elucidate the deletion subtypes of 88 subjects. In our cohort, 32 had a Type 1 and 49 had a Type 2 deletion. The remaining seven subjects had unique or atypical deletions that were either smaller (n=5) or larger (n=2) than typically described and were further characterized by array-based comparative genome hybridization. In two subjects both the PWS region (15q11.2) and the newly described 15q13.3 microdeletion syndrome region were deleted. The subjects with a unique or an atypical deletion revealed distinct phenotypic features. In conclusion, unique or atypical deletions were found in ∼8% of the deletion subjects with PWS in our cohort. These novel deletions provide further insight into the potential role of several of the genes within the 15q11.2 and the 15q13.3 regions.
Similar content being viewed by others
Introduction
Prader–Willi syndrome (PWS; OMIM #176270) is an imprinted neurobehavioral condition affecting many organ systems. Major manifestations include early childhood obesity, hyperphagia, hypotonia with poor suck and poor weight gain in infancy, early childhood obesity, mild to moderate mental retardation, hypogonadism, growth hormone insufficiency causing short stature, characteristic facial appearance, characteristic neurobehavior and frequently psychiatric disturbance including abnormal restricted repetitive behavior (eg, skin picking, obsession, compulsion, sameness behavior, etc.).1, 2
PWS is caused by an absence of a functionally active paternal contribution in the chromosome 15q11.2–q13 region, in contrast to Angelman syndrome (AS) that is due to an absence of a maternal contribution in the same chromosomal region. PWS occurs via three distinct genetic mechanisms: deletion of paternally expressed genes in 15q11.2 (65–75%), maternal uniparental disomy (UPD; 20–30%) and imprinting defect (ID; 1–3%).1, 3, 4
Deletions in PWS and AS are subdivided into two main subgroups (Type 1 and 2) and the breakpoints (BPs) are flanked by low-copy repeats (LCR). The common BP regions are proximally at BP1 or BP2, and distally at BP3. Both Type 1 and 2 deletions are almost always de novo events. For the purpose of this study, we used the designation of Type 1 deletion to encompass the region between BP1 and BP3 (∼6 Mb in size), and Type 2 deletion to encompass the region from BP2 to BP3 (∼5.3 Mb in size).5, 6, 7 However, there are rare PWS and AS patients with an atypical distal BP at BP4 or BP5.8, 9 Additionally, in a few patients, the 15q11.2-q13 region may be deleted as a result of an unbalanced translocation, which will yield unique BPs within proximal 15q.10, 11
Despite PWS being a well-characterized genetic disorder, specific genetic factors contributing to each of the distinct clinical features of PWS are not clearly understood.1, 4 Increasing evidence suggest that the loss of the paternally expressed snoRNA gene cluster, SNORD116, has a significant role in many of the features of the PWS phenotype.12 It is unclear whether the four, non-imprinted genes (NIPA1, NIPA2, CYFIP1 and GCP5) localized to the interval between BP1 and BP213 contribute towards the PWS phenotype. Non-PWS individuals with just a small deletion between BP1 and BP2 have a variable phenotype. For instance, some individuals apparently have developmental delay and/or autistic features, but some of the parents have the same deletion without apparent effect.14
Several studies have investigated phenotypic characteristics between PWS individuals with Type 1 vs Type 2 deletions, but there has been a lack of consensus among the different studies. For example, Butler et al15 reported individuals with Type 1 deletion (n=12) showed worse adaptive behavior, more severe compulsive behavior and more impairments in reading, math skills and visual perception than those with Type 2 deletion (n=14). In contrast, Dykens and Roof16 (Type 1, n=26; Type 2, n=29) and Milner et al17 (Type 1, n=14; Type 2, n=32) did not find significant phenotypic differences between the two main deletion subtypes. One problem is that none of these studies had a large sample size in each of the two deletion subtypes (size varied from 12 to 32), or used the same measures for the phenotypic assessments. Furthermore, not all of the studies utilized methods that would fine-map the distal BP. Thus, if some of the patients had distal BPs more proximal or distal than BP3, this would affect the genes involved in the deletion and potentially the phenotype.
As the genotype–phenotype relationships become clearer, it will be clinically important to readily subtype the deletion class. Current standard of clinical care in the determination of the deletion class of PWS involves use of fluorescent in situ hybridization (FISH) typically using only the SNRPN probe in the 15q11.2-q13 region. If the FISH test is positive for a deletion, and DNA methylation analysis reveals an absence of the paternally inherited 15q11.2 region, then few clinical geneticists at this time will delve any further to determine the size of deletion.
Methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA) is a relatively new technique to detect changes of copy number and DNA methylation status, and has been used successfully in PWS and AS diagnosis.18, 19 Compared with the traditional diagnostic methods, such as chromosomal and FISH analysis, MS-MLPA provides more detailed information on the size of the deletion as well as distinguishing PWS from AS.
The aims of the current study were twofold. One aim was to evaluate the utility of the MS-MLPA technique to delineate the size of the deletion in our cohort of 88 PWS subjects with a deletion. Another aim was to perform genotype–phenotype correlations on the seven subjects who had either a unique deletion (ie, having one or two novel BPs) or an atypical deletion (not a Type 1 or 2 deletion, but having a distal BP in another LCR/duplicon like BP5).
Materials and methods
Sample
We obtained DNA samples from 88 PWS individuals with a deletion whose subtypes were unknown (see Supplementary Table S1 for details). In addition, we have examined an additional 145 DNA samples to compare the DNA methylation patterns across the diagnostic categories. These include 16 PWS subjects with maternal UPD, 5 PWS subjects with an ID, 8 AS subjects with a deletion, 1 AS subject with paternal UPD, 2 AS subjects with an ID, 4 family members with a known maternal duplication of the 15q11.2-q13,20 1 subject with maternal isodicentric duplication of chromosome 15 (47,XX,+inv dup(15)(pter-q11::q11-pter)), 105 subjects with a history of early-onset morbid obesity (EMO) and 4 phenotypically normal control subjects. The research protocol and informed consents were approved by the University of Florida Institutional Review Board.
MS-MLPA
A commercial MS-MLPA kit (ME028, MRC-Holland, Amsterdam, The Netherlands) for PWS/AS was utilized. Two versions were created for ME028 (versions A1 and B1, see Supplementary Table S2 for details). Depending on the version, this kit contains 25–32 probes specific for sequence along the length of the 15q11.2-q13 region to detect copy number changes in genes and other important areas. We used the A1 kit for all our deletion subjects. Some of our subjects were also run on the B1 kit. The manufacturer's protocol was followed for the DNA preparation, ligation, enzyme digestion and multiplex PCR reaction. After the final PCR step, the Applied Biosystems 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA) and Peak Scanner Software (version 1.0; Applied Biosystems) were used for capillary electrophoresis and fragment analysis. We utilized the MLPA interpretation protocol developed by Drs Karin Buiting and Bernhard Horsthemke (personal communication) for determination of copy number changes, and manufacturer's protocol to determine the DNA methylation changes. All MS-MLPA results were reviewed independently by two investigators (SJK and DJD). Whenever the results were inconclusive, the MS-MLPA reaction was repeated.
The array-based comparative genome hybridization
MS-MLPA results from a subset of 35 PWS subjects were verified by an enhanced version of a chromosome 15 specific genomic microarray. These 35 subjects include the 7 subjects with unique or atypical deletions identified by MS-MLPA, and 28 subjects who were study participants recruited as part of our Rare Disease Clinical Research Network center grant. The array-based comparative genome hybridization (aCGH) included a total of over 38 000 oligos across 15q, with a higher density of oligos spanning ∼10 Mb of the 15q11.2-q14 interval at the Baylor College of Medicine (Agilent Technologies Inc., Santa Clara, CA, USA).7
Clinical evaluation
The clinical evaluations for the subjects with a unique or an atypical deletions were done during clinic visits and/or during clinical research encounters.
Results
Copy number analyses
Among 88 PWS subjects with deletion, we identified 32 subjects (36.4%) with Type 1 deletion and 49 subjects (55.7%) with Type 2 deletion (Supplementary Table S1). The remaining seven PWS subjects (8.0%) had unique or atypical deletions other than Type 1 or 2 (Figure 1). Among eight AS subjects with deletion, five subjects (62.5%) had Type 1 deletion and the remaining three subjects (37.5%) had Type 2 deletion. We also observed microdeletions within SNRPN in one of five PWS subjects with an ID and two of two AS subjects with an ID. We incidentally identified two microdeletions among 105 EMO subjects (one with a deletion of the 15q26 region and the other with a deletion between BP1 and BP2). The four subjects with an interstitial duplication of the 15q11.2-q13 region of maternal origin, as well as the subject with maternal isodicentric duplication of proximal 15q showed increased dosage of the respective chromosomal intervals consistent with their molecular diagnoses.
Seven unique or atypical PWS deletions. The position of genes and genetic markers (circles) in the chromosomal 15q11.2-q14 region are shown. In the PWS region (shown in blue), there are six paternal-only expressed unique copy genes (MKRN3, MAGEL2, NECDIN, C15orf2 and SNURF-SNRPN and a family of six snoRNA gene clusters). Only UBE3A and ATP10A (shown in red), related to AS, have maternal-only expression in mouse and humans, and this imprinted expression is limited to certain tissue-specific regions (ie, mostly regions in the brain). The bipartite imprinting center (IC) lies proximal to SNURF-SNRPN and within the 3 Mb PWS/AS imprinted region. The cluster of GABA receptor genes (GABRB3, GABRA5 and GABRG3), OCA2 (Type 2 albinism) and HERC2 are not imprinted and have biparental expression (shown in open black circle). The jagged vertical lines denote the common PWS deletion BPs, which lie within the segmental duplications associated with BP1–BP5. Type 1 deletions extend from BP1 to BP3 and Type 2 deletions extend from BP2 to BP3. The seven unique or atypical deletions (ie, neither Type 1 or 2) identified from this study are shown in solid lines with base pair positions of BPs confirmed by aCGH. These base pair positions are derived from the UCSC genome browser March 2006 (hg18) freeze (http://www.genome.ucsc.edu/). There is a lack of agreement in the literature regarding the order of the genes between BP1 and BP2. Note that there are more copies of the SNORD116 and SNORD115 snoRNA genes than are shown, and map distances are not drawn exactly to scale.
DNA methylation analyses
DNA methylation analyses utilizing the NDN and SNRPN probes confirmed characteristic DNA methylation patterns for PWS, AS and normal control subjects (Table 1).
Agreement rate between the MS-MLPA and aCGH
The agreement rate between MS-MLPA and aCGH was 100%. However, the aCGH did provide a more precise location of the BPs.
Case studies
The clinical and research records of the seven subjects with a unique or an atypical deletion confirmed by both MS-MLPA and aCGH were reviewed in more detail (Figure 1 and Table 2). A complete description of each case is in the Supplementary Data. An abbreviated description is given below for each case. The deletions and translocation described in these seven cases were all de novo in origin.
Case 1 PW235P
MS-MLPA showed a deletion of MKRN3 to intron 2 of ATP10A, sparing exons 1 and 2. aCGH showed a unique deletion of ∼2.46 Mb. The features not typical of PWS in this subject include the macrocephaly,21 large hands and feet, tall stature for PWS (all before the start of growth hormone), large birth weight, normal skin pigmentation and lower-than-average pain tolerance (Table 2).
Case 2 HBTB-113
The subject came to our attention after his brain was donated to the Human Brain Tissue Bank (HBTB) program at the University of Florida. Therefore, we obtained minimal records on him including his autopsy and his hospital records before his death. MS-MLPA found a unique deletion between BP1 and a site telomeric to ATP10A sparing the GABRB3 and more distal genes. These results were confirmed by aCGH, which showed a deletion size of ∼3.60 Mb (Table 2).
Case 3 PW173P
A clinical karyotype analysis revealed an unbalanced karyotype interpreted as 45,XY,der(6)t(6;15)(p25;q12),-15. FISH analysis confirmed a 15q11.2 deletion with the SNRPN probe. MS-MLPA revealed a deletion of NIPA1 to GABRB3 while sparing OCA2 (there were no MS-MLPA probes for GABRA5 and GABRG3). The research aCGH demonstrated a unique deletion extending from the pericentromeric 15 region to intron 3 of GABRG3, sparing exons and genes telomeric to intron 3 including OCA2. A clinical aCGH demonstrated no loss of unique 6p material. The size of the deletion was ∼5.09 Mb. Atypical features for PWS in this subject include microcephaly, normal pigmentation, lack of skin picking, and a pleasant, mild-mannered personality with lack of argumentative behavior (Table 2).
Case 4 PW246P
A clinical karyotype and FISH analyses revealed 45,XX,der(14),t(14;15)(p11.1;q13).
MS-MLPA revealed a deletion of NIPA1 to GABRB3 while sparing OCA2. The research aCGH revealed a unique deletion extending from the pericentromeric 15 region to intron 5 of GABRG3. The size of the deletion was ∼5.58 Mb. A clinical aCGH demonstrated no loss of unique 14q material. Features not typical of PWS include microcephaly, lower-than-average birth weight for PWS,22, 23 and later onset of obesity. She lacked the typical PWS facial gestalt. In addition, she was reportedly functioning at a higher level than typical for PWS, according to the experienced group home staff.
Case 5 PW133P
MS-MLPA revealed a deletion of the gene interval from BP1 to within the OCA2 gene. The aCGH revealed a unique distal BP that was within intron 18 of OCA2, sparing exons 1–18 of OCA2. The features not typical of PWS in this patient were lower-than-average birth weight for PWS.22
Case 6 PW231P
MS-MLPA revealed a deletion from MKRN3 to APBA2. The aCGH demonstrated a deletion of ∼9.06 Mb extending from (and including) MKRN3 through CHRNA7. Therefore, this subject had an atypical deletion involving BP2 proximally and BP5 distally. The features not typical of PWS include milder hypotonia and feeding difficulties as a neonate, less delay of developmental milestones, macrocephaly (even before the start of growth hormone treatment), frontal bossing, inverted nipples, bilateral transverse palmar creases, large hands and feet, absence of seizures and higher-than-typical IQ for PWS.
Case 7 PW259P
MS-MLPA detected a deletion from MRKN3 to APBA2. The aCGH demonstrated a 9.31 Mb deletion extending from MKRN3 through DKFZp434L187, ∼250 kb telomeric of CHRNA7. Like case 6, this subject had a BP2–BP5 atypical deletion. The features not typical of PWS include lower-than-typical birth weight, the much more severe developmental delay, the microcephaly, atypical facial features, increased hypotonia and the prolonged failure-to-thrive.
Discussion
A deletion in the paternal 15q11.2-q13 chromosomal region is the most common cause of PWS; however, it has become clear that Type 1 and 2 deletions are not the only type of deletions that occur in individuals with PWS. Among our cohort of 88 deletion subjects with PWS, we found 36.4% with Type 1 deletion, 55.7% with Type 2 deletion and 8% with a unique or an atypical deletion. The higher number of Type 2 vs Type 1 is similar to the findings of other studies.15, 16, 17 The prevalence of unique or atypical deletions was higher than what we expected and has not previously been reported for PWS, but was comparable to the prevalence rate (9.1%) found in AS.7
The 15q11.2-q13 region is highly vulnerable to structural rearrangements, such as deletions, duplications, supernumerary marker chromosomes, and translocations due to presence of LCRs in the region.24, 25 In our subjects with unique or atypical deletion sizes, two subjects (cases 6 and 7) had both proximal and distal BPs involving LCRs (ie, BP2 and BP5). Another three of our subjects (cases 1, 2 and 5) had a proximal BP in an LCR (ie, either BP1 or BP2), but the distal BP was unique and located proximal to BP3 in an interval between ATP10A and OCA2. Cases 3 and 4 had an unbalanced translocation with neither BP involving an LCR in the 15q11.2-q13.3 region.
In this study, we also examined genotype–phenotype relationships in our seven subjects with unique or atypical deletion sizes. Three of our subjects (cases 1, 3 and 4) with an intact OCA2 gene had normal skin pigmentation comparable to other members of their family. This is not surprising given the role of OCA2 in skin pigmentation.26 Case 2 came to our attention only after his brain was donated to our brain bank, so we cannot comment on his skin pigmentation. The clinic notes we obtained have no mention of his pigmentation. Case 5 had a deletion within intron 18 of OCA2 and was hypopigmented. Case 6 and 7 had large deletions that included OCA2, but hypopigmentation was not noted. In our clinics, we have found that hypopigmentation in PWS deletion cases is much harder to discern in the more deeply pigmented ethnic and racial groups. Case 6 is Hispanic and case 7 is Black, which probably explains why hypopigmentation was not appreciated in these two subjects.
One subject had a much smaller deletion (2.46 Mb) than a typical deletion (Case 1). His deletion spared the GABAA receptor subunit gene clusters and OCA2. Interestingly, he had an above average birth weight, macrocephaly, large hands/feet and tall stature for PWS (before he started growth hormone), lower-than-average pain tolerance, and somewhat delayed onset of the voracious appetite and food-stealing behavior. These atypical features contributed to the delay in his PWS diagnosis. However, the moderate skin picking behavior in this subject was rather puzzling. Given the postulated role of GABA in the mediation and perception of pain,27 we would have anticipated that sparing the GABAA receptor subunit gene clusters would have lessened this behavior. In addition, he manifested the common behavioral characteristics of individuals with PWS including temper tantrums, easily angered and rigidness. Furthermore, his cognitive IQ was 63, well within the typical range for PWS.28 Unfortunately, we do not have detailed behavioral information on case 2 whose deletion also spared the GABAA receptor subunit gene clusters. Therefore, based only on this one subject, it would appear that sparing the GABAA receptor subunit gene clusters does not alter the typical PWS neurobehavior.
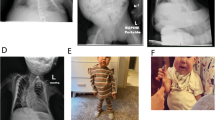
Interestingly our two subjects (cases 6 and 7) with large deletions spanning >9 Mb between BP2 and BP5 had certain similarities and differences in their phenotypic presentation. In both the CHRNA7 gene was deleted, but case 7 had an additional 250 kb deletion telomeric to CHRNA7 involving the DKFZp434L187 transcript. CHRNA7 encodes the alpha-7 subunit of the neuronal nicotinic acetylcholine receptor.29 Recently several groups have implicated CHRNA7 as a candidate gene for the 15q13.3 microdeletion syndrome whose clinical manifestations include facial and digital dysmorphology, expressive language deficit, and various neuropsychiatric disorders, such as schizophrenia, autism, epilepsy and mental retardation.30, 31, 32, 33 The finding that cases 6 and 7 had large deletions encompassing the 15q13.3 region prompted us to request an EEG be done on both of them. Similar to those with the 15q13.3 microdeletion, case 6 had both clinical and EEG-documented seizure activities. Surprisingly, her parents, teachers and health care providers were unaware of her absence seizures until her abnormal seizure related behavior (eg, looking away, lip smacking, etc.) correlated with the EEG wave changes. Case 7 also had an abnormal EEG; however no clinical seizure activity has been noted in this subject. As he is still young, it would be reasonable to assume that he is at increased risk for seizure activities. In addition, both case 6 and 7 lacked the typical facial features of PWS, but did share some facial features in common with the 15q13.3 microdeletion subjects (Figure 2).
The two PWS subjects with large atypical deletions and lacking the typical PWS facial gestalt. Case 6 at 9 months (a),  years (b) and 7 years (c, d). At 9 months (a) she appears well nourished with a weight for length at the 75th percentile. Features include a depressed and wide nasal bridge, blunted columella, round face and long, smooth philtrum. Case 7 at 13 months (e, f), 24 months (g) and 4 years (h). Note the thin, frail appearance with G-tube in place as an infant with a weight for length at <3rd percentile (e, f). Weight began to increase at 24 months. Facial features include a prominent metopic suture, round face, depressed and wide nasal bridge, blunted columella, and short and smooth philtrum.
years (b) and 7 years (c, d). At 9 months (a) she appears well nourished with a weight for length at the 75th percentile. Features include a depressed and wide nasal bridge, blunted columella, round face and long, smooth philtrum. Case 7 at 13 months (e, f), 24 months (g) and 4 years (h). Note the thin, frail appearance with G-tube in place as an infant with a weight for length at <3rd percentile (e, f). Weight began to increase at 24 months. Facial features include a prominent metopic suture, round face, depressed and wide nasal bridge, blunted columella, and short and smooth philtrum.
Although sharing some similarities, case 6 and 7 did have drastic differences in their clinical manifestations. Case 6 had a much milder postnatal course, less severe developmental delay, macrocephaly, and higher intellectual and academic functioning. At 7.2 years she had a cognitive IQ score of 89 and an achievement score of 109 both of which are significantly higher than typically found in PWS.28 This is surprising as her deletion encompasses both the PWS region as well as the 15q13.3 microdeletion region. In contrast, case 7 had a much more severe failure-to-thrive phase requiring a feeding tube for 2.5 years, microcephaly, markedly delayed gross and fine motor skills as well as speech development. At 4.9 years he was still not walking independently. Case 7's clinical findings are more in line with what we would have expected from deleting two microdeletion syndrome regions. At this point it is not clear why there is such a dramatic difference between cases 6 and 7 who share a similar-sized deletion of ∼9 Mb. There are several possibilities for the differences. First, the genetic variation in the 15q13.3 region on the maternal chromosome may become ‘unmasked’ by the paternally derived deletion, which may have contributed to the more severe clinical phenotype in case 7. Second, genetic variation elsewhere in the genome may have contributed to the composite clinical features in case 7. A great deal of variability in clinical features has been described for individuals with the 15q13.3 microdeletion including some parents with the deletion who lack the clinical features found in their offspring.33 Furthermore, the 15q13.3 region, like the 22q11.2 and 16p12.1 regions, may be another example of a ‘susceptibility region’ that requires a ‘second hit’ elsewhere in the genome to become fully penetrant as a neurodevelopmental phenotype.34, 35 Third, the DKFZp434L187 transcript (deleted in case 7) downstream to CHRNA7 may require further evaluation for its potential role in the more severe developmental delay and failure-to-thrive, although this is very unlikely given that this transcript has not been shown to code for any important function to our knowledge.
We identified several strengths and weaknesses of the MS-MLPA technique. MS-MLPA reliably detected the deletion as evidenced by 100% agreement with our laboratory data and aCGH. It also provided more detailed information regarding the size of the deletion than standard FISH due to many (25–32) probes across the 15q11.2-q13 region. Thus, the MS-MLPA technique allows for a reasonable approximation of the BPs and deletion size. Compared with aCGH, the MS-MLPA technique was much more labor/cost-effective, although aCGH provides more precise information regarding the extent of the deletion. In addition, the DNA methylation component of MS-MLPA allows differentiation between PWS and AS deletions, and maternal and paternal 15q11.2 duplications, as well as between uniparental and biparental disomy. Therefore, a compelling argument could be made that MS-MLPA should be the first test used when contemplating AS or PWS as a possible diagnosis, especially since important genotype–phenotype correlations will likely be forthcoming.
In this study, we had an opportunity to evaluate both the A1 and B1 versions of the commercial MS-MLPA kit. The A1 version has two probes for OCA2, which are useful to identify unique deletions with distal BPs between GABRB3 and BP3. If we had not used the A1 version, we would have missed the unique deletion sizes in cases 3, 4 and 5. The B1 version, on the other hand, is designed to provide additional coverage at the bipartite AS and PWS imprinting center and SNORD116 regions. Taken together, the A1 version appears superior in detecting deletion sizes of AS and PWS, whereas the B1 version is superior in identifying small deletions in the imprinting center and/or the SNORD116 region.
In conclusion, this study makes several important points. First, deletions should be characterized by accurately determining both their proximal and distal BPs, rather than just their proximal BP. The designation of Type 1 deletion should be strictly reserved for those deletions between BP1 and BP3, and Type 2 deletion for those deletions between BP2 and BP3. This is important for genotype–phenotype counseling in the future, as unique or atypical (ie, not type 1 or 2) deletions due to a novel distal and/or proximal BP could result in a milder or more severe phenotype than either Type 1 or Type 2 deletions. This may also contribute to the lack of current consensus in genotype–phenotype comparisons of Type 1 vs Type 2 deletions, as the distal BPs were not always well delineated in many of these studies. Second, the frequency of deletions that are neither Type 1 nor 2 (8% in our study) is higher than previously recognized in PWS. Third, genotype–phenotype studies on individuals with unique or atypical deletions have the potential to elucidate the role of the various genes in the 15q11.2 region. Fourth, the mechanisms for producing unique deletions need further clarification as at least five of the subjects in this study had at least one BP that did not occur at a 15q11.2-q13 LCR region. Fifth, the differences in clinical manifestations of our two subjects with a deletion extending into the 15q13.3 region further underscore the clinical variability of the 15q13.3 microdeletion syndrome. In our future studies, we plan to obtain more detailed phenotypic information using specific neurobehavioral measures, such as cognitive and behavioral profiles, comorbid psychiatric illnesses, and response to certain medication, to more fully examine genotype–phenotype relationships in these subjects.
References
Cassidy SB, Driscoll DJ : Prader-Willi syndrome. Eur J Hum Genet 2009; 17: 3–13.
Dykens EM, Cassidy SB, King BH : Maladaptive behavior differences in Prader-Willi syndrome due to paternal deletion versus maternal uniparental disomy. Am J Ment Retard 1999; 104: 67–77.
Horsthemke B, Buiting K : Genomic imprinting and imprinting defects in humans. Adv Genet 2008; 61: 225–246.
Nicholls RD, Knepper JL : Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu Rev Genomics Hum Genet 2001; 2: 153–175.
Christian SL, Bhatt NK, Martin SA et al: Integrated YAC contig map of the Prader-Willi/Angelman region on chromosome 15q11-q13 with average STS spacing of 35 kb. Genome Res 1998; 8: 146–157.
Butler MG, Fischer W, Kibiryeva N, Bittel DC : Array comparative genomic hybridization (aCGH) analysis in Prader-Willi syndrome. Am J Med Genet A 2008; 146: 854–860.
Sahoo T, Bacino CA, German JR et al: Identification of novel deletions of 15q11q13 in Angelman syndrome by array-CGH: molecular characterization and genotype-phenotype correlations. Eur J Hum Genet 2007; 15: 943–949.
Wang NJ, Liu D, Parokonny AS, Schanen NC : High-resolution molecular characterization of 15q11-q13 rearrangements by array comparative genomic hybridization (array CGH) with detection of gene dosage. Am J Hum Genet 2004; 75: 267–281.
Sahoo T, Shaw CA, Young AS et al: Array-based comparative genomic hybridization analysis of recurrent chromosome 15q rearrangements. Am J Med Genet A 2005; 139: 106–113.
Buiting K, Horsthemke B, Prader-Willi Syndrome Association: Molecular Genetic Findings in Prader-Willi Syndrome. in Merlin G Butler, PDKL, Barbara Y Whitman (eds): Management of Prader-Willi Syndrome, 3rd edn, Birkhäuser: Springer, 2006, pp 58–73.
Mignon-Ravix C, Depetris D, Luciani JJ et al: Recurrent rearrangements in the proximal 15q11-q14 region: a new breakpoint cluster specific to unbalanced translocations. Eur J Hum Genet 2007; 15: 432–440.
Duker AL, Ballif BC, Bawle EV et al: Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome. Eur J Hum Genet 2010; 18: 1196–1201.
Chai JH, Locke DP, Greally JM et al: Identification of four highly conserved genes between breakpoint hotspots BP1 and BP2 of the Prader-Willi/Angelman syndromes deletion region that have undergone evolutionary transposition mediated by flanking duplicons. Am J Hum Genet 2003; 73: 898–925.
Burnside RD, Pasion R, Mikhail FM et al: Microdeletion/microduplication of proximal 15q11.2 between BP1 and BP2: a susceptibility region for neurological dysfunction including developmental and language delay. Hum Genet 2011.
Butler MG, Bittel DC, Kibiryeva N, Talebizadeh Z, Thompson T : Behavioral differences among subjects with Prader-Willi syndrome and type I or type II deletion and maternal disomy. Pediatrics 2004; 113: 565–573.
Dykens EM, Roof E : Behavior in Prader-Willi syndrome: relationship to genetic subtypes and age. J Child Psychol Psychiatry 2008; 49: 1001–1008.
Milner KM, Craig EE, Thompson RJ et al: Prader-Willi syndrome: intellectual abilities and behavioural features by genetic subtype. J Child Psychol Psychiatry 2005; 46: 1089–1096.
Procter M, Chou LS, Tang W, Jama M, Mao R : Molecular diagnosis of Prader-Willi and Angelman syndromes by methylation-specific melting analysis and methylation-specific multiplex ligation-dependent probe amplification. Clin Chem 2006; 52: 1276–1283.
Bittel DC, Kibiryeva N, Butler MG : Methylation-specific multiplex ligation-dependent probe amplification analysis of subjects with chromosome 15 abnormalities. Genet Test 2007; 11: 467–475.
Boyar FZ, Whitney MM, Lossie AC et al: A family with a grand-maternally derived interstitial duplication of proximal 15q. Clin Genet 2001; 60: 421–430.
Nellhaus G : Head circumference from birth to eighteen years. Practical composite international and interracial graphs. Pediatrics 1968; 41: 106–114.
Butler MG, Sturich J, Myers SE, Gold JA, Kimonis V, Driscoll DJ : Is gestation in Prader-Willi syndrome affected by the genetic subtype? J Assist Reprod Genet 2009; 26: 461–466.
Miller JL, Lynn CH, Driscoll DC et al: Nutritional Phases in Prader-Willi Syndrome. Am J Med Genet 2011; in: press.
Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH : Large genomic duplicons map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region (15q11-q13). Hum Mol Genet 1999; 8: 1025–1037.
Donlon TA, Lalande M, Wyman A, Bruns G, Latt SA : Isolation of molecular probes associated with the chromosome 15 instability in the Prader-Willi syndrome. Proc Natl Acad Sci USA 1986; 83: 4408–4412.
Spritz RA, Bailin T, Nicholls RD et al: Hypopigmentation in the Prader-Willi syndrome correlates with P gene deletion but not with haplotype of the hemizygous P allele. Am J Med Genet 1997; 71: 57–62.
Enna SJ, McCarson KE : The role of GABA in the mediation and perception of pain. Adv Pharmacol 2006; 54: 1–27.
Miller J, Kranzler J, Liu Y et al: Neurocognitive findings in Prader-Willi syndrome and early-onset morbid obesity. J Pediatr 2006; 149: 192–198.
Agulhon C, Abitbol M, Bertrand D, Malafosse A : Localization of mRNA for CHRNA7 in human fetal brain. Neuroreport 1999; 10: 2223–2227.
Helbig I, Mefford HC, Sharp AJ et al: 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet 2009; 41: 160–162.
van Bon BW, Mefford HC, Menten B et al: Further delineation of the 15q13 microdeletion and duplication syndromes: a clinical spectrum varying from non-pathogenic to a severe outcome. J Med Genet 2009; 46: 511–523.
Sharp AJ, Mefford HC, Li K et al: A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet 2008; 40: 322–328.
Ben-Shachar S, Lanpher B, German JR et al: Microdeletion 15q13.3: a locus with incomplete penetrance for autism, mental retardation, and psychiatric disorders. J Med Genet 2009; 46: 382–388.
Girirajan S, Eichler EE : Phenotypic variability and genetic susceptibility to genomic disorders. Hum Mol Genet 2010; 19: R176–R187.
Antonacci F, Kidd JM, Marques-Bonet T et al: A large and complex structural polymorphism at 16p12.1 underlies microdeletion disease risk. Nat Genet 2010; 42: 745–750.
Acknowledgements
We gratefully acknowledge the participation and provision of information provided by the families at the University of Florida and in the Rare Disease Natural History Study. We thank Krista Garner and Christy Lynn for their expert assistance in clinical assessments, Drs Karin Buiting and Bernhard Horsthemke for sharing their MLPA protocol, and Dr Robert Nicholls for critical comments on parts of this manuscript. This work was supported in part by the Hayward Foundation (DJD), the National Institutes of Health (NIH) U54 HD061222 and RR019478 (SJK; JLM; ALB; TXS; DJD), and the NIH/National Center for Research Resources CTSA Grant 1UL 1RR029890. SJK is supported in part by 2007 NARSAD young investigator award, 2008 PWSA (USA) Research Award, NIH R03MH083673 and NIH K23MH082883 awards.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Kim, SJ., Miller, J., Kuipers, P. et al. Unique and atypical deletions in Prader–Willi syndrome reveal distinct phenotypes. Eur J Hum Genet 20, 283–290 (2012). https://doi.org/10.1038/ejhg.2011.187
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2011.187
Keywords
This article is cited by
-
Genetic subtypes and phenotypic characteristics of 110 patients with Prader-Willi syndrome
Italian Journal of Pediatrics (2022)
-
Recommendations for the diagnosis and management of childhood Prader-Willi syndrome in China
Orphanet Journal of Rare Diseases (2022)
-
First report of two successive deletions on chromosome 15q13 cytogenetic bands in a boy and girl: additional data to 15q13.3 syndrome with a report of high IQ patient
Molecular Cytogenetics (2019)
-
Gastro-oesophageal reflux - an important causative factor of severe tooth wear in Prader-Willi syndrome?
Orphanet Journal of Rare Diseases (2018)
-
Diagnoses and characteristics of autism spectrum disorders in children with Prader-Willi syndrome
Journal of Neurodevelopmental Disorders (2017)