Abstract
A mosaicism is defined by the presence of two or more populations of cells with different genotypes in one individual. Chromosomal germinal mosaicism occurs in germ cells before the onset of meiosis. Previously, few studies have described germinal mosaicism. In this study, we report on two siblings who carried identical pure and direct interstitial 4q22.2q32.3 duplication. Procedure investigations included complete clinical description, conventional cytogenetic analysis, fluorescence in situ hybridization (FISH), comparative genomic hybridization (CGH) array experiments and microsatellite study searching for parental origin of the duplication. Microarray CGH and further FISH experiments with BAC clones showed the same 70.8 Mb direct duplication, dup(4)(q22.2q32.3). Molecular studies of the 4q duplication were consistent with maternal origin associated with mitotic or meiotic rearrangements. This structural chromosomal aberration was associated in both cases with increased nuchal translucency, growth retardation and dysmorphy. Cardiopathy and lung malformations were only evident in the first case. These clinical manifestations are similar to those previously reported in previous studies involving pure 4q trisomy of the same region, except for thumb and renal abnormalities that were not obvious in the presented cases. The amplified region included genes involved in neurological development (NEUROG2, MAB21L2, PCDH10/18 and GRIA2). The recurrent 4q duplication in these siblings is consistent with a maternal ovarian germinal mosaicism. This is the first description of germinal mosaicism for a large chromosomal duplication and highlights that genetic counselling for apparently de novo chromosome aberration should be undertaken with care.
Similar content being viewed by others
Introduction
A mosaicism is defined by the presence of two or more cell lines with different genotypes in one individual, who has developed from a single fertilized egg.1 Different types of mosaicism exist, such as germinal mosaicism (restricted to germ cells) or tissue mosaicism. The most common form of mosaicism detected through prenatal diagnosis involves trisomies. Chromosomal germinal mosaicism is rare and occurs in early germ cells (oogonia or spermatogonia) before the onset of meiosis.1 These mitotic errors can be nondisjunctions (abnormality of chromosome number) or the production of structural rearrangements (deletion, duplication, inversion, insertion or translocation). Germinal mosaicism becomes evident when two siblings are born with the same de novo chromosomal abnormality. Germinal mosaicism is independent of maternal age.2 Few studies in literature have described germinal mosaicism corresponding essentially to aneuploidies and structural chromosomal aberrations including mostly isochromosomes and deletions3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19 (for review, see Röthlisberger and Kotzot20).
In this study, we report two siblings carrying the same pure and direct 4q22.2q32.3 duplication characterized using microarray comparative genomic hybridization (CGH) and due to ovarian germinal mosaicism.
Materials and methods
Clinical report
The first pregnancy of a 24-year-old woman was marked by increased nuchal translucency (4.5 mm) on first-trimester ultrasound examination. Parents were nonconsanguineous and both familial histories were unremarkable. Chorionic villi were sampled for chromosomal analysis that showed a large long arm on chromosome 4. This chromosomal abnormality was confirmed on amniotic fluid analysis at 16 weeks. After genetic counselling and according to the French law, the pregnancy was terminated at 20 weeks of gestation. The female foetus was hypotrophic. Weight, height, head circumference and biparietal diameter measurements were 221.6 g (<5th percentile), 15 cm (5th percentile), 25 cm (25th percentile) and 14 cm (<25th percentile), respectively. Physical examination showed cranio-facial dysmorphism with elongated skull, large nose, long philtrum, large mouth with self-effacing cupidon arc, badly hemmed ears and neck oedema (Supplementary Figure 1). Internal examination revealed pulmonary lobulation defect with two lobes in the right lung and a single one in the left lung. Interauricular communication was also noted. The placenta was hypotrophic (86.8 g versus approximately 160 g for normal controls at the same term) but with normal aspect.
During the second pregnancy, increased nuchal translucency was again detected (5 mm) on first-trimester ultrasound examination. Chorionic villi were sampled and chromosomal analysis showed an abnormal long arm on chromosome 4 similar to that observed in the first foetus. Regular foetus ultrasound examinations showed normal amniotic fluid volume and no malformations were detected in the foetus whose growth was normal. After several discussions, both parents decided to continue the pregnancy. At 38 weeks of gestation, a boy was vaginally delivered. His weight was 3040 g (<25th percentile). Physical examination showed general hypotonia and facial dysmorphism with narrow and horizontal palpebral fissures, prominent nose and moderate microretrognatia. At 6 weeks of age, his weight, height and head circumference were 4400 g (<25th percentile), 55.5 cm (<50th percentile) and 37.5 cm (<25th percentile), respectively. The smile response was acquired, axial tonus was correct and no other abnormality was specified. Facial dysmorphic features were stable. At the age of 6 months, the boy had no specific medical treatment. His weight, height and head circumference were 7070 g (−1 SD), 64 cm (−1 SD) and 42 cm (−1 SD), respectively. He was able to catch close objects but not distant ones. He was able to burst into laughter. The seating position was not completely acquired and axial hypotonia was still present. At the age of 1 year, his weight, height and head circumference were 8900 g (−1 SD), 70 cm (−1 SD) and 46 cm (−1 SD), respectively. Axial hypotonia was still marked, resulting in mild kyphosis when seating and in the absence of any possibility to stand up. Psychomotricity support was begun. At the age of 32 months, his height, weight and head circumference were 80 cm (−3.5 SD), 10,500 kg (−2.5 SD) and 49 cm (−1 SD), respectively. The child was unable to walk alone and still needed some help. He was able to walk on all fours limbs and to stand up against a wall or a table. His language was very poor, limited to two or three disyllabic words, whereas hearing explorations proved normal. Facial dysmorphism was unchanged (Supplementary Figure 2). Educational support was continued.
Conventional cytogenetic analysis
Chromosome analyses were performed from uncultured and cultured trophoblast cells, amniotic cells and peripheral lymphocytes using standard procedures (RHG banding, CBG banding and a high-resolution banding technique obtained after cell culture synchronization and BrdU incorporation).
DNA extraction
Genomic DNA was isolated from peripheral blood (parent's propositus), placenta (case 1) and cultured trophoblast cells (case 2) using the Qiagen DNeasy Blood and Tissue Kit (Qiagen, Courtaboeuf, France). Extracted DNA concentrations were estimated using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Extracted DNAs were used for array CGH (aCGH) and microsatellite analysis.
Array comparative genomic hybridization (aCGH)
The genomic imbalances of placenta and trophoblast cells were analysed by aCGH using 105K oligonucleotide arrays (Hu-105A, Agilent Technologies, Massy, France). All array hybridizations were performed according to the manufacturer's recommended protocols. In brief, 3 μg of genomic DNA was digested with AluI (5 units) and RsaI (5 units) for 2 h at 37 °C and fluorescently labelled with the Agilent Genomic DNA labelling kit PLUS (Agilent Technologies). A male or female human genomic DNA (Promega, Charbonnière, France) was used as reference. Experiments were conducted in dye-swap. Cy5-dUTP patient DNA and its gender-matched reference labelled with Cy3-dUTP were denatured and preannealed with Cot-1 DNA and Agilent blocking reagent before hybridization for 40 h at 20 r.p.m. in a 65 °C rotating hybridization oven (Agilent Technologies). After washing, the slides were scanned on an Agilent Microarray Scanner. Captured images were processed with Feature Extraction 9.1 (Agilent Technologies) software and data analysis was performed with CGH Analytics 3.5 (Agilent Technologies). Copy number variations (CNVs) were considered significant if they were defined by three or more oligonucleotides spanning at least 50 Kb and contained at least one gene and were not identified in the Database of Genomic Variants.
Fluorescence in situ hybridization (FISH)
FISH analyses were performed on trophoblast cells and metaphase spreads from both parental and propositus lymphocytes. The whole-chromosome painting (WCP) probe specific for chromosome 4 was used according to the manufacturer's recommendations (Amplitech, Compiègne, France).
BAC clones specific for the 4q chromosomal region (RP11-397E7 located at 4q21.3, RP11-79M20 located at 4q22.1, RP11-451M10 located at 4q22.1, RP11-16I17 located at 4q22.3, RP11-81J9 located at 4q25, RP11-501E13 located at 4q25, RP11-77P11 located at 4q28.1, RP11-481K16 located at 4q31.21, RP11-301H24 located at 4q31.21, RP11-655B23 located at 4q31.23, RP11-177L7 located at 4q32.3 and RP11-18D7 located at 4q35) were used. BAC DNAs were labelled by nick translation using a FITC-dUTP nucleotide or Rhodamine-dUTP nucleotide (Roche Diagnostics, Rungis, France).
Microsatellite analysis
Thirteen polymorphic markers from chromosome 4 (see location and characteristics in Table 1) were coamplified by multiplex polymerase chain reaction (PCR). Primer sequences were designed according to the GeneDB locus description from The Wellcome Trust Sanger Institute. Multiplex PCR was carried out following standard protocols using 50 ng of DNA in 50 μl reaction volume and fluorescence dye-labelled primers (10 pmol each primer, 10 mM Tris-HCl (pH 9), 50 mM KCl, 2.25 mM MgCl2, 0.2 mM each dNTP and 2 U Taq Polymerase (Taq Core Kit; MP Biomedicals, Illkirch, France)). PCR conditions were denaturation at 95 °C for 5 min, followed by 20 cycles of 95 °C for 40 s, 58 °C for 40 s, 72 °C for 40 s and a final 72 °C extension step for 7 min (Veriti Thermal Cycler; Applied Biosystems, Courtaboeuf, France). PCR products were then separated onto an ABI Prism 3130 analyser (Applied Biosystems), with the GeneScan 500LIZ as size standard. Data were analysed using GeneMapper 4.0 software (Applied Biosystems).
Results
Conventional cytogenetic analysis
In case 1, a direct analysis of trophoblast cells showed an abnormal female 46,XX,add(4) karyotype with an additional chromosomal region located on 4q. In case 2, cytogenetic analyses on trophoblast cells revealed the same abnormal chromosome 4 with an additional region at the long arm (46,XY,add(4)) (Figure 1a). In both cases, this was interpreted as representing either a duplication of the long arm 4q or another rearrangement such as a translocation or an insertion. Analyses of parents' peripheral blood lymphocytes showed normal standard karyotypes.
R-banding partial karyotype of chromosome 4 from trophoblast cells showing additional material on the long arm of one chromosome 4 (right) (a). Chromosome 4 DNA dye-swap profile from array-based CGH analysis showing gain (duplication) for oligonucleotides located in the 4q22.2q32.3 region for case 1 (b) and case 2 (c).
DNA microarray assay
CGH analysis showed in both cases a gain of the 4q22.2q32.3 region (case 1 in Figure 1b and case 2 in Figure 1c). For both cases, the analysis revealed a proximal break point located on 4q22.2 (position 94 128 982) and a distal break point on 4q32.3 (position 164 941 617). These genomic positions were determined using version 18 of the human genome built (http://genome.ucsc.edu/). Thus, a 70.8 Mb interstitial region was amplified. Analyses revealed other variations (gain or loss) on chromosome 4 and on other chromosomes. These changes corresponded to CNVs previously reported in the database of genomic variants (http://projects.tcag.ca/variation/).
FISH analysis
The WCP probe specific for chromosome 4 showed complete hybridization on normal chromosome 4 and on the abnormal chromosome 4 (Supplementary Figure 3A). No hybridization signal was detected on any other chromosome. This finding excluded the possibility of an insertion or a translocation. FISH experiments could not be realized in case 1 because only uncultured placenta was available.
In case 2, BACs RP11-16I17 (4q22.3), RP11-81J9 (4q25), RP11-77P11 (4q28.1), RP11-481K16 (4q31.21) and RP11-655B23 (4q31.23) gave one signal on normal chromosome 4 and two signals on duplicated chromosome 4. No hybridization signal of these BAC probes was detected on any other chromosome. Moreover BACs RP11-451M10 (4q22.1), RP11-177L7 (4q32.3) and RP11-18D7 (4q35) showed one signal on both normal chromosome 4 and the duplicated one. Thus, we confirmed that the proximal break point was located between BACs RP11-451M10 (4q22.1) and RP11-16I17 (4q22.3) and the distal one between BACs RP11-655B23 (4q31.23) and RP11-177L7 (4q32.3). These results were in accordance with the propositus CGH array profile.
To clearly define the nature of the chromosomal rearrangement, double colour hybridization using BAC probes RP11-501E13 (4q25) and RP11-301H24 (4q31.21) was realized. The position of hybridization signals on the duplicated chromosome showed that the rearrangement was a direct duplication (Supplementary Figure 3B). Schematic representation of the position of BAC probes is summarized in Supplementary Figure 3C.
To exclude low mosaicism for the chromosome 4 duplication, FISH analyses using BACs RP11-301H24 (4q31.21) were performed in parent cells. Two signals were observed in 200 interphase nuclei and 100 metaphases for each parent, thus excluding duplication 4q mosaicism in lymphocytes.
Microsatellite analysis
Microsatellite marker analysis was informative for parental origin determination and also in specifying the mechanism of the duplication rearrangement (Table 2). Indeed, D4S427, D4S422, D4S1579 and D4S3334 marker profiles were consistent with maternal origin for both cases (Table 2). As a representative result, D4S3334 analysis is shown in Figure 2a. Moreover, D4S1573, D4S191, D4S3024, D4S427, D4S194 and D4S1579 marker analyses indicated that both duplicated maternal alleles were different for case 1 and were identical for case 2 (Table 2). These results pointed out an interchromosomal rearrangement for case 1 and an intrachromosomal one for case 2. As a representative result, D4S194 analysis is shown in Figure 2b. A possible segregation of these informative microsatellite markers illustrating this recombination is represented in Figure 3.
In summary, these conventional and molecular cytogenetic experiments allowed a precise characterization of the chromosomal formula of both siblings. We concluded that both cases carried a pure and direct dup(4)(q22.2q32.3) of maternal origin. As the maternal karyotype was normal, these results suggested an ovarian germinal mosaicism.
Discussion
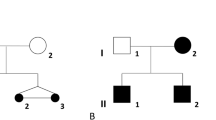
We report on two siblings (a female foetus and a 3-year-old male infant) carrying the same structural abnormality with normal parental karyotypes. In both cases, prenatal ultrasound had detected increased nuchal translucency. Both presented facial dysmorphic elements such as long philtrum, large mouth with self-effacing cupidon arc, low-set and badly hemmed ears, horizontal palpebral fissures, prominent nose, moderate microretrognatia and microcephaly. They also showed prenatal and/or postnatal growth retardation. These phenotypes were associated with a de novo direct dup(4)(q22.2q32.3). In both cases, CGH array indicated a 70.8 Mb duplication size and microsatellite analysis showed that the duplication had maternal origin. Microsatellite analysis also specified that the duplication resulted from an interchromosomal rearrangement in the first case and from an intrachromosomal one in the second case. Thus, these results suggest an ovarian 46,XX/46,XX,dup(4)(q22.2q32.3) mosaicism.
Germinal mosaicism is independent of maternal age,2 and becomes evident when two siblings are born with the same de novo chromosomal abnormality. Numerous studies described germinal mosaicism involving molecular defects such as gene deletion, gene mutation or intragenic intron rearrangements (for review, see Zlotogora1). Cases of germinal mosaicism involving structural and numeric chromosomal rearrangements are rare but described (for review, see Röthlisberger and Kotzot20). Chromosomal germinal mosaicism is because of mitotic errors before the onset of meiosis. These errors could be nondisjunctions (abnormality of chromosome number) or the production of structural rearrangements (deletion, duplication, inversion, insertion or translocation). Few studies have highlighted parental germinal mosaicism with an abnormal number of chromosomes such as trisomy 18 and 213, 4, 5, 6, 7 or monosomy X.3, 8 Until now, only 11 studies have described germinal mosaicism involving structural chromosomal aberration.9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19 Indeed, five studies reported structural abnormality with a maternal mosaicism origin.9, 10, 11, 18, 19 Engel et al9 reported a familial pseudodicentric chromosome (5;21) occurring on maternal germline mosaicism as revealed by microsatellite marker analysis. Another study described sibling cases with a subtelomeric 5.8 Mb deletion on chromosome 15 as a result of maternal germinal mosaicism10 using microarray CGH and polymorphic marker analysis. A recurrent case of chromosome 18 inversion [46,XY,+18,der(18;inv(18))(q10;q10)] was also described as a result of a maternal germinal mosaic.11 Finally, two recurrent deletions [del(16)(q11.3q12.2) and del(22)(q13.3qtel)] were reported.18, 19 With regard to male mosaics, another study reported sibling propositus with chromosome 14 structural abnormalities as a consequence of father testicular mosaicism,12 and two more cases of recurrent paternal deletions on chromosomes 11 and 13 were described.16, 17 Our report is the first to describe a large duplication rearrangement due to germinal mosaicism. Chromosomal regions around break points did not contain segmental duplications (UCSC Genome Browser database search) that could favour the duplication mechanism. In this study, six informative microsatellite analyses indicated the occurrence of an allelic crossover during maternal gamete production.
Two hypotheses can be established to explain case 1 interchromosomal recombination and case 2 intrachromosomal recombination (Figure 4). First, allelic crossover could have occurred in the prophase of the first meiotic division (Figure 4a). Indeed, theorical meiotic recombination frequency in the duplicated region having a size of 70.8 Mb is approximately 70%.21 Second, allelic crossover could have occurred during oogonia mitotic division (Figure 4b). However, the first hypothesis is more plausible. In both situations, four gamete types could be produced in ovaries to explain our observations. These allelic recombinations in the duplicated 4q region between siblings could explain the phenotype differences observed between both siblings by affecting, for example, gene regulation.
To our knowledge, 17 studies have described the pure 4q duplication region covering the 4q22q32 region22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38 (total reports included the 4q21q35.2 chromosomal region analysed by conventional cytogenetic and FISH experiments, see Supplementary Table 1). No prenatal case has ever been reported.
Comparing clinical features of the 22 patients including ours (Supplementary Table 1) showed the occurrence of growth or psychomotor/mental retardation (15 out of 22 reported cases), microcephaly (11 of 22), dysmorphic elements including epicanthus folds (15 of 22), prominent nasal bridge (16 of 22) and low-set ears (14 of 22). Short philtrum (9 of 22) and micrognathia (8 of 22) are relatively common. It has been suggested that thumb and renal abnormalities could be associated with the 4q22q23 and/or the 4q25q31.3 region.30, 31 Contrary to the reports of these researchers, we did not observe thumb or renal abnormalities in our patients having a 4q22q32 duplication. To our knowledge, the present study is the first report of a pure and recurrent 4q duplication investigated with CGH microarray technology and microsatellite analysis. CGH on chromosomes has been used in the study by Elghezal et al.32 CGH array analysis allowed a more precise description of distal and proximal break points because of a better resolution of 15 kb (average probe spacing). Gene analysis using the UCSC Genome Browser database (http://genome.ucsc.edu/) of CGH microarray results listed more than 110 duplicated genes. Thus, it was difficult to correlate observed phenotypes with implicated genes. Nevertheless, several duplicated genes expressed in neurogenesis, such as neurogenin 2 (NEUROG2, 4q25),39 mab-21-like 2 (MAB21L2, 4q31),40 protocadherin 10/18 (PCDH10, 4q28.3 and PCDH18, 4q31) and glutamate receptor ionotropic AMPA2 (GRIA2, 4q32q33),41 could be implicated in the phenotype. In particular, an overexpression of these genes could explain the general hypotonia and mental delay observed in the second patient.
In conclusion, this report highlighted that genetic counselling for apparently de novo chromosome aberration should be undertaken with care with regard to germinal mosaicism. The actual risk for de novo structural aberration taking into account the abortion risk of prenatal diagnostic is estimated at approximately 0.5–1%.20
References
Zlotogora J : Germ line mosaicism. Hum Genet 1998; 102: 381–386.
Delhanty JD : Mechanisms of aneuploidy induction in human oogenesis and early embryogenesis. Cytogenet Genome Res 2005; 111: 237–244.
Uehara S, Yaegashi N, Maeda T et al: Risk of recurrence of fetal chromosomal aberrations: analysis of trisomy 21, trisomy 18, trisomy 13, and 45,X in 1076 Japanese mothers. J Obstet Gynaecol Res 1999; 25: 373–379.
Bruyere H, Rupps R, Kuchinka BD, Friedman JM, Robinson WP : Recurrent trisomy 21 in a couple with a child presenting trisomy 21 mosaicism and maternal uniparental disomy for chromosome 21 in the euploid cell line. Am J Med Genet 2000; 94: 35–41.
Sachs ES, Jahoda MG, Los FJ, Pijpers L, Wladimiroff JW : Trisomy 21 mosaicism in gonads with unexpectedly high recurrence risks. Am J Med Genet Suppl 1990; 7: 186–188.
Mehes K, Kosztolanyi G : A possible mosaic form of delayed centromere separation and aneuploidy. Hum Genet 1992; 88: 477–478.
Conn CM, Cozzi J, Harper JC, Winston RM, Delhanty JD : Preimplantation genetic diagnosis for couples at high risk of Down syndrome pregnancy owing to parental translocation or mosaicism. J Med Genet 1999; 36: 45–50.
Beaulieu Bergeron M, Tran-Thanh D, Fournet JC, Lemyre E, Lemieux N, Bouron-Dal Soglio D : Male pseudohermaphroditism and gonadal mosaicism in a 47,XY,+22 fetus. Am J Med Genet A 2006; 140: 1768–1772.
Engel U, Bohlander SK, Bink K, Hinney B, Laccone F, Bartels I : Pseudo dicentric chromosome (5;21): a rare example of maternal germline mosaicism. Hum Reprod 2001; 16: 63–66.
Rump P, Dijkhuizen T, Sikkema-Raddatz B et al: Drayer's syndrome of mental retardation, microcephaly, short stature and absent phalanges is caused by a recurrent deletion of chromosome 15(q26.2-->qter). Clin Genet 2008; 74: 455–462.
Prabhakara K, Wyandt HE, Huang XL, Prasad KS, Ramadevi AR : Recurrent proximal 18p monosomy and 18q trisomy in a family with a maternal pericentric inversion of chromosome 18. Ann Genet 2004; 47: 297–303.
Masada CT, Olney AH, Fordyce R, Sanger WG : Partial deletion of 14q and partial duplication of 14q in sibs: testicular mosaicism for t(14q;14q) as a common mechanism. Am J Med Genet 1989; 34: 528–534.
Hajianpour A, Murer-Orlando M, Docherty Z : Germ line mosaicism for chromosome 5 Cri-du-chat deletion? Am J Hum Genet 1991; 49: 217.
Gardner RJ, Dockery HE, Fitzgerald PH et al: Mosaicism with a normal cell line and an autosomal structural rearrangement. J Med Genet 1994; 31: 108–114.
Chilcote RR, Le Beau MM, Dampier C et al: Association of red cell spherocytosis with deletion of the short arm of chromosome 8. Blood 1987; 69: 156–159.
Grossfeld PD, Mattina T, Lai Z et al: The 11q terminal deletion disorder: a prospective study of 110 cases. Am J Med Genet Part A 2004; 129A: 51–61.
Brandriff B, Gordon LA, Crawford BB et al: Sperm chromosome analysis to assess potential germ cell mosaicism. Clin Genet 1988; 34: 85–89.
Hoo JJ, Lowry RB, Lin CC, Haslam RH : Recurrent de novo interstitial deletion of 16q in two mentally retarded sisters. Clin Genet 1985; 27: 420–425.
Tabolacci E, Zollino M, Lecce R et al: Two brothers with 22q13.3 deletion syndrome and features suggestive of the Clark-Baraitser syndrome phenotype. Clin Dysmorphol 2005; 14: 127–132.
Röthlisberger B, Kotzot D : Recurrence risk in de novo structural chromosomal rearrangements. Am J Med Genet A 2007; 143A: 1708–1714.
Morton NE, Lindsten J, Iselius L, Yee S : Data and theory for a revised chiasma map of man. Hum Genet 1982; 62: 266–270.
Vogel W, Siebers JW, Gunkel J : [Phenotypic variation in partial trisomy 4q (author's transl)]. Humangenetik 1975; 28: 103–112.
Dutrillaux B, Laurent C, Forabosco A et al: Partial 4q trisomy. Apropos of 3 cases. Ann Genet 1975; 18: 21–27.
Taylor KM, Francke U, Brown MG, George DL, Kaufhold M : Inverted tandem (‘mirror’) duplications in human chromosomes: -nv dup 8p, 4q, 22q. Am J Med Genet 1977; 1: 3–19.
Fryns JP, van den Berghe H : Partial duplication of the long arm of chromosome 4. Ann Genet 1980; 23: 52–53.
Halal F, Vekemans M, Chitayat D : Interstitial tandem direct duplication of the long arm of chromosome 4 (q23-q27) and possible assignment of the structural gene encoding human aspartylglucosaminidase to this segment. Am J Med Genet 1991; 39: 418–421.
Jeziorowska A, Ciesla W, Houck Jr GE et al: Cytogenetic and molecular identification of a de novo direct duplication of the long arm of chromosome 4(q21.3-->q31.3). Am J Med Genet 1993; 46: 83–87.
Navarro EG, Romero MC, Exposito IL et al: De novo interstitial tandem duplication of chromosome 4(q21-q28). Am J Med Genet 1996; 62: 297–299.
Goodman BK, Capone GT, Hennessey J, Thomas GH : Familial tandem duplication of bands q31.1 to q32.3 on chromosome 4 with mild phenotypic effect. Am J Med Genet 1997; 73: 119–124.
Muraki K, Katano R, Hiraki Y, Ueda K, Fujita H : A case of an interstitial tandem direct duplication of long arm of chromosome 4: 46, XY, dup (4) (q25q31.3) de novo. Hiroshima J Med Sci 1997; 46: 105–108.
Maltby EL, Barnes IC, Bennett CP : Duplication involving band 4q32 with minimal clinical effect. Am J Med Genet 1999; 83: 431.
Elghezal H, Sendi HS, Monastiri K et al: Large duplication 4q25-q34 with mild clinical effect. Ann Genet 2004; 47: 419–422.
Lin S, Kirk EP, McKenzie F, Francis C, Shalhoub C, Turner AM : De novo interstitial duplication 4(q28.1q35) associated with choanal atresia. J Paediatr Child Health 2004; 40: 401–403.
Hubert E, Sawicka A, Wasilewska E, Midro AT : Partial trisomy of long arm of chromosome 4 as a result of dir dup (4)(q27q31.3) de novo. Genet Couns 2006; 17: 211–218.
Otsuka T, Fujinaka H, Imamura M, Tanaka Y, Hayakawa H, Tomizawa S : Duplication of chromosome 4q: renal pathology of two siblings. Am J Med Genet A 2005; 134: 330–333.
Cernakova I, Kvasnicova M, Lovasova Z et al: A duplication dup(4)(q28q35.2) de novo in a newborn. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 2006; 150: 113–116.
ECARUCA: Database ID3940 (http://agserver01.azn.nl:8080/ecaruca/ecaruca.jsp).
DECIPHER: Database ID251350 (https://decipher.sanger.ac.uk/application/).
Ribes V, Stutzmann F, Bianchetti L, Guillemot F, Dolle P, Le Roux I : Combinatorial signalling controls Neurogenin2 expression at the onset of spinal neurogenesis. Dev Biol 2008; 321: 470–481.
Wong RL, Chow KL : Depletion of Mab21l1 and Mab21l2 messages in mouse embryo arrests axial turning, and impairs notochord and neural tube differentiation. Teratology 2002; 65: 70–77.
Narisawa-Saito M, Iwakura Y, Kawamura M et al: Brain-derived neurotrophic factor regulates surface expression of alpha-amino-3-hydroxy-5-methyl-4-isoxazoleproprionic acid receptors by enhancing the N-ethylmaleimide-sensitive factor/GluR2 interaction in developing neocortical neurons. J Biol Chem 2002; 277: 40901–40910.
Acknowledgements
We thank C Metay, A Briand and the technical team for help with CGH array experiments and results interpretation. We thank L Cuisset for microsatellite analysis and A-E Mas for case 1 photography. We also thank the DHOS (Direction de l'Hospitalisation et de l'Organisation des Soins) for their support in the development of array CGH platform (Paris Sud).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Tosca, L., Brisset, S., Petit, F. et al. Recurrent 70.8 Mb 4q22.2q32.3 duplication due to ovarian germinal mosaicism. Eur J Hum Genet 18, 882–888 (2010). https://doi.org/10.1038/ejhg.2010.46
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2010.46
Keywords
This article is cited by
-
A case of germline mosaicism for a 7q32.1q33 deletion in a sperm donor: consequences on pregnancy follow-up and recommendations
Basic and Clinical Andrology (2020)
-
Somatic/gonadal mosaicism for structural autosomal rearrangements: female predominance among carriers of gonadal mosaicism for unbalanced rearrangements
Molecular Cytogenetics (2016)