Abstract
We studied a man with distal hypospadias, partial anomalous pulmonary venous return, mild limb-length inequality and a balanced translocation involving chromosomes 9 and 13. To gain insight into the etiology of his birth defects, we mapped the translocation breakpoints by high-resolution comparative genomic hybridization (CGH), using chromosome 9- and 13-specific tiling arrays to analyze genetic material from a spontaneously aborted fetus with unbalanced segregation of the translocation. The chromosome 13 breakpoint was ∼400 kb away from the nearest gene, but the chromosome 9 breakpoint fell within an intron of Basonuclin 2 (BNC2), a gene that encodes an evolutionarily conserved nuclear zinc-finger protein. The BNC2/Bnc2 gene is abundantly expressed in developing mouse and human periurethral tissues. In all, 6 of 48 unrelated subjects with distal hypospadias had nine novel nonsynonymous substitutions in BNC2, five of which were computationally predicted to be deleterious. In comparison, two of 23 controls with normal penile urethra morphology, each had a novel nonsynonymous substitution in BNC2, one of which was predicted to be deleterious. Bnc2−/− mice of both sexes displayed a high frequency of distal urethral defects; heterozygotes showed similar defects with reduced penetrance. The association of BNC2 disruption with distal urethral defects and the gene's expression pattern indicate that it functions in urethral development.
Similar content being viewed by others
Introduction
Hypospadias, when the urethral orifice is located on the ventral aspect of the penis, is found in 0.7% of live births.1 Although previous studies have suggested that the majority of cases are sporadic and multifactorial,2 multiple families have been described with isolated hypospadias and apparent autosomal dominant inheritance.3 Genes involved in the etiology of hypospadias and their interactions with environmental factors were recently reviewed.4 Androgen signaling is crucial to normal penile development, and most known genetic causes of hypospadias involve defects in androgen signaling. For instance, mutations in the 5-alpha reductase gene (SRD5A2) or the androgen receptor gene (AR) may cause severe hypospadias, usually in conjunction with other genital malformations.5, 6, 7 Additionally, mutations in MAMLD1 cause a rare X-linked form of the disorder.8 MAMLD1 contains an SF1 binding site and transactivates the noncanonical Notch target gene Hes3 to augment testosterone production in Leydig cells.9 Other genes that are involved in penile or urethral patterning have been linked to human hypospadias, including HOXA13, mutations of which cause hand-foot-genital syndrome.10, 11
Basonuclin 2 (BNC2) is a gene of unknown function that is evolutionarily conserved in vertebrates.12 The predicted BNC2 protein is about 1100 amino acids in length and contains a nuclear localization signal and three zinc-finger motifs,13, 14 with a similar overall structure to Basonuclin 1 (BNC1), a protein found in keratinocytes15 and germ cells16 that acts as a transcription factor for ribosomal RNA17, 18 and possibly also RNA polymerase II-dependent genes.19 BNC1 and BNC2 show nonoverlapping patterns of expression, and BNC2 but not BNC1 localizes to nuclear speckles, suggesting that BNC2 functions in mRNA processing.20 Bnc2−/− knockout mice die perinatally, with cleft palate and other craniofacial abnormalities.21
We serendipitously identified a man with a de novo balanced 9;13 translocation and distal hypospadias, partial anomalous pulmonary venous return (PAPVR) and mild lower-limb length inequality. We used a novel array comparative genomic hybridization (CGH) approach to characterize the translocation breakpoints, taking advantage of the availability of genetic material from a spontaneously aborted fetus he fathered with unbalanced segregation of the translocation. The CGH results allowed us to rapidly determine that BNC2 is a candidate gene for PAPVR and hypospadias. To corroborate the role of BNC2 in these birth defects, we screened additional subjects with APVR or hypospadias for BNC2-coding mutations and carefully analyzed the phenotype of Bnc2-knockout mice.
Materials and methods
Oligonucleotide array CGH
Genomic DNA was purified from cultured cells from products of conception (POC) by standard methods. CGH was performed by Nimblegen Systems Inc. (Madison, WI, USA) using chromosome-specific tiling arrays containing 385 000 isothermal 50–75-mer oligonucleotide probes and pooled normal human male reference DNA (Promega, Madison, WI, USA). The mean probe density was one probe per 255 bp for chromosome 9 (Catalog No. B3740001-00-01) and one probe per 225 bp for chromosome 13 (Catalog No. B3744001-00-01).
Fluorescence in situ hybridization (FISH)
Metaphase chromosomes were prepared from either PHA-stimulated whole-blood lymphocytes or EBV-immortalized lymphoblasts. BAC clones were purchased from BACPAC resources (Oakland, CA, USA). BAC DNAs were isolated using the BACMAX DNA isolation kit (Epicentre, Madison, WI, USA), labeled with Spectrum Orange according to the manufacturer's instructions (Vysis, Downers Grove, IL, USA), precipitated, resuspended in LSI/WCP hybridization buffer (Vysis), and hybridized to slides overnight at 37 °C. Washed and dehydrated slides were counterstained with DAPI II (Vysis) and visualized with an Olympus BX-61 fluorescence microscope. Images were captured with a CCD camera and Cytovision software (Applied Imaging, San Jose, CA, USA).
Quantitative RT-PCR
Multiple human fetal tissue RNAs were purchased from Clontech Laboratories (Mountain View, CA, USA, Catalog No. 636747). Lymphoblastoid cell RNAs were isolated using Tripure (Roche, Indianapolis, IN, USA), and reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA) according to the manufacturers’ protocols. Transcripts of BNC2, LMO7 and a control gene (cyclophilin A) were measured by real-time PCR using TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA, USA) Hs00214187_m1 (BNC2 exons 5 and 6), Hs00417700_m1 (BNC2 exons 3 and 4), Hs00245600_m1 (LMO7) and cyclophilin A (Catalog No. 4310883E). The BNC2 and LMO7 probes contained a 6-carboxy-fluorescein phosphoramidite (FAM dye) label at the 5′-end and were designed to hybridize across intron/exon junctions. The cyclophilin A control probe was labeled with VIC dye and used in multiplex with the BNC2 or LMO7 probes. We used cDNA derived from human skeletal muscle RNA (Clontech Laboratories) for calibration and generation of standard curves. Reactions were performed in triplicate. Samples with replicate coefficient of variation >20% were repeated.
Allelic expression
The 5′-untranslated region (UTR) of LMO7 was amplified with primers 5′-TCAGATAATCTTAAGGCTGTTGG-3′ and 5′-TTTCCTCATGAGCAGTGTGC-3′. The 5′-UTR of BNC2 was amplified with primers 5′-CTTTTCAAAGTTGCTGAAATAAAA-3′ and 5′-TTGCATTTAATGGCCTCAGA-3′. Genomic PCR products from the proband were sequenced and a heterozygous SNP identified in each gene (LMO7: rs9565177; BNC2: rs1999032). RT-PCR products were amplified from DNAse-treated cDNA isolated from lymphoblastoid cells carrying the balanced translocation as described above, and sequenced. Control reactions omitting reverse transcriptase were performed to rule out amplification of contaminating genomic DNA. Electropherograms of genomic and cDNA sequences were compared to determine whether both alleles of LMO7 and BNC2 were expressed.
Mutation screening
Informed consent/assent for this study was obtained from human subjects under protocols approved by the University of Texas Southwestern Medical Center and the University of Utah Institutional Review Boards. Genomic DNA was extracted from whole blood of the proband and forty other subjects with surgically-repaired APVR. BNC2 exons 1, 2, 2a, 3, 4, 5 and 6 (numbered according to ref.12) were PCR-amplified using primers designed with Primer-3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3) and both strands sequenced with the same primers. Primer sequences are available upon request. Sequence data were analyzed using Seqman (DNAStar, Madison, WI, USA). BNC2 coding exons from subjects with hypospadias and controls with normal penile urethral anatomy were analyzed in a similar manner by Polymorphic DNA Technologies, Inc. (Alameda, CA, USA). Missense substitutions were analyzed by SIFT22 using the web site http://sift.jcvi.org/.
Mouse studies
Animal studies were approved by the Université Paris Descartes Animal Use Committee. For X-gal staining of sections, newborns were snap frozen in OCT (Miles, Elkhart, IN, USA) and isopentane, and cut with a cryomicrotome at 7 μ M. Sections were then fixed in formaldehyde/glutaraldehyde (2%:0.2%), incubated in the presence of X-gal overnight at 37 °C and counterstained with hematoxylin/eosin. Genotype and sex were determined by PCR.
Results
The proband (Supplementary Figure 1) was a healthy 33 year old man with unrepaired mild coronal hypospadias and PAPVR that required surgical repair at age 23. The right upper and right middle pulmonary veins drained anomalously to the superior vena cava. He also had mild lower-limb length inequality treated with a shoe lift, but no other medical problems. His spouse suffered two miscarriages; cytogenetic study of the POC from the second miscarriage revealed an unbalanced 9;13 translocation, karyotype 46,XX,der(9)t(9;13)(p22;q22). Further testing showed that the proband carried a de novo balanced 9;13 translocation (Supplementary Figure 1) that was not transmitted to his healthy daughter.
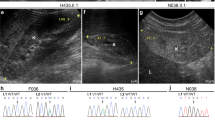
We assumed that the molecular chromosomal breakpoints were the same in the proband with the balanced translocation as in the POC with the unbalanced translocation. Taking advantage of the availability of POC cells, we performed tiling array CGH using chromosome 9- and 13-specific oligonucleotide arrays in order to fine-map the breakpoints. The CGH results confirmed that the POC was trisomic for 13q22-qter (Figure 1a) and monosomic for 9pter-p22 (Figure 1b). Breakpoints were mapped from the CGH data to a 3 kb interval of chromosome 9 (chr9:16 508 500–16 511 500, GRCh37/hg19 genome assembly) and a 4 kb interval of chromosome 13 (chr13:76 839 999–76 843 999).
Array CGH and FISH mapping of translocation breakpoints for chromosomes 13 (a) and 9 (b). CGH graphs show coordinates from NCBI 36.1 (hg18) genome assembly (X axis); log2 relative hybridization signal intensity versus control (Y axis). Arrows on metaphase spreads point to hybridization signals (red).
Both breakpoints were confirmed using FISH with chromosome 9 and 13 BAC clones. Both clones consistently showed two signals in the expected locations when hybridized to normal control metaphases. In contrast, each probe consistently showed three signals, one with normal intensity and two with reduced intensity, when hybridized to the proband's metaphases (Figure 1). The FISH results confirmed that the breakpoints were the same in the proband and the POC, and ruled out a sizable cryptic deletion at either junction.
The chromosome 13 breakpoint did not disrupt any known gene. It was ∼400 kb downstream of the nearest gene, LMO7, which encodes a putative transcriptional regulator containing PDZ and LIM protein–protein interaction domains. The chromosome 9 breakpoint fell within a large intron of the BNC2, which encodes a zinc-finger protein gene of unknown function.14 BNC2 shows a highly complex pattern of alternative splicing, promoters and polyadenylation sites, and encodes a plethora of predicted proteins, some of which share no amino acids with each other.12 However, nearly all BNC2 transcripts contain exons 5 and 6 (numbered according to ref. 12). Both of these exons are required for the function of the protein as they encode all the important features of the protein including the six zinc fingers and the nuclear localization signal. As the breakpoint disrupts BNC2 between exons 4 and 5, it is predicted to cause a null mutation. No BNC2 coding mutations were identified in the proband, whose phenotype thus appeared likely to result from BNC2 haploinsufficiency.
The tissue distribution of LMO7 mRNA and two major BNC2 transcripts was assessed by quantitative RT-PCR using human adult and fetal multiple tissue RNA panels (Supplementary Figure 2). In adult tissues, BNC2 was especially prevalent in the uterus, with expression also observed in kidney, lung, prostate, testes and trachea. The highest levels of LMO7 were observed in heart, lung and skeletal muscle. In fetal tissues, BNC2 transcripts containing exons 3 and 4 were highly expressed in both heart and penis; transcripts containing exons 5 and 6 were also enriched in fetal penis, whereas LMO7 also showed the highest levels in fetal heart and penis.
To determine if the translocation perturbed either LMO7 or BNC2 expression, we identified a heterozygous SNP in the 5′-UTR of each gene. RT-PCR products from lymphoblastoid cells carrying the balanced translocation were heterozygous for these SNPs, with approximately equal peak heights (Figure 2a), similar to that of sequences of genomic PCR products. These data indicate that, at least in lymphoblastoid cell lines, LMO7 or BNC2 alleles on both the normal and the derivative chromosomes were transcribed at roughly equal levels, although presumably the BNC2 transcript(s) from the derivative-9 chromosome lack exons 5 and 6. Additionally, the LMO7 mRNA level in immortalized lymphoblasts from the proband was within the range demonstrated by the normal controls (Figure 2b). Although we were able to amplify trace amounts of biallelic BNC2 transcripts in lymphoblastoid cells from the proband, the level of expression in this cell type was not sufficient for quantitation.
To corroborate a role for BNC2 in birth defects, we sequenced the major BNC2 coding exons (1, 2, 2a, 3–6; ref.12) in the proband and 40 unrelated subjects with surgically repaired APVR (Table 1). None of the males in this sample had hypospadias. Two novel nonsynonymous substitutions were found in exon 5: D760E was found in a subject with partial APVR and an ASD (APVR7), and L414V was found in a subject with total APVR (APRV36). The aspartic acid residue in the first substitution is evolutionarily conserved among BNC2 orthologs but not between BNC2 and its paralog, BNC1.14 This residue is not within any known functional protein domain. The second substitution alters a leucine that is conserved among BNC2 orthologs and its BNC1 paralog, but also does not fall within any known functional domains. This subject's parents, both without known medical conditions, were studied and his mother was found to carry the same substitution. An additional novel nonsynonymous substitution, A923V, was found in exon 6 in three subjects, one with PAPVR and two with TAPVR. It changes an alanine residue conserved among BNC2 orthologs, but not between BNC2 and BNC1, and is also not in any known domains. None of the nonsynonymous substitutions in APVR subjects were predicted to be deleterious by SIFT.22
We next screened for BNC2 coding variations in 48 subjects with distal hypospadias vs 23 controls with normal penile urethra anatomy. The results (Table 1) showed eight heterozygous novel nonsynonymous substitutions in six hypospadias subjects (one subject had three nonsynonymous substitutions) vs two novel nonsynonymous substitutions in two controls (6/48 (12.5%) vs 2/23 (8.7%), P=NS). Four nonsynonymous substitutions in three hypospadias subjects were predicted to be deleterious by SIFT, but so was one of the nonsynonymous substitutions present in a control. Two of the substitutions in hypospadias subjects, L414V and A923V, were also found in APVR subjects.
We then analyzed Bnc2 expression in the penis and urethra of bnc2+/− and bnc2−/− mice.21 These mice carry a lacZ gene inserted into a large Bnc2 intron and express β-galactosidase under the control of the Bnc2 promoter. Frozen sections of male bnc2+/− newborns were stained for β-gal activity using X-gal as a chromogenic substrate. Transverse sections of the entire mouse in the lower abdominal region showed that bnc2–lacZ expression is highest in the developing penis (Figure 3a). β-gal activity is specific to the cells of the preputial lamella and in a zone of mesenchymal cells just exterior to the corpus spongiosum and just exterior to the condensed mesenchyme of the future os penis (Figure 3b). As homozygous mutant mice die soon after birth, examination was limited to newborns, a high proportion of which showed the presence of a cleft in the ventral part of the external genitalia in both males and females (Figure 4a). This was presumably because of the inability of the urethral folds to fuse at the ventral midline. Male phallic size was normal in both genotypes. Heterozygous mice also showed a urethral defect, but with reduced penetrance and severity compared with bnc2−/− mice (Figure 4a; Table 2). Dye injections into the bladder demonstrated patency of the urethra, which terminated as a hypospadic urethral meatus in homozygotes (Figure 4b). No obvious cardiac defects were seen in these mice, and the developing heart and surrounding vasculature did not clearly stain for β-gal activity.
Urethral defects in bnc2+/− and bnc2−/− mice. (a) Gross appearance of external genitalia in newborn males and females (ventral view). In both male and female heterozygous newborns, a narrow epithelialized furrow because of incomplete closure of the urethral folds is visible. In the homozygous mutant the furrow is hypospadic, with a wider and deeper groove than the hets and with a pit-like depression at the distal end. (b) Dye injection into the bladders of newborn males reveals defects in the penile urethral meatus of homozygotes.
Discussion
Disease-associated balanced translocations are powerful tools to identify developmental genes.23 Although the availability of the human genome sequence and BAC clones for any region of interest have greatly facilitated mapping studies, characterizing a translocation breakpoint beyond the limit of resolution of FISH remains laborious. In this report, we demonstrate how array CGH, another technological innovation enabled by the human genome sequence, can be used to fine-map translocation breakpoints rapidly. The application of CGH is limited to situations where there is a gain or loss of genetic material, which is not the case for truly balanced translocations. However, when genetic material from an unbalanced segregant is available, as in the present case, CGH can map breakpoints from the unbalanced translocation that can then be readily verified by FISH studies of the balanced translocation. We know of only one other case where an unbalanced meiotic segregant was used to map a constitutional balanced translocation breakpoint.24
We studied a man with a de novo balanced 9;13 translocation, distal hypospadias, PAPVR and mild limb-length inequality. The chromosome 9 breakpoint fell in an intron of BNC2, a large (∼470 kb) gene of unknown function with a highly complex pattern of alternative transcriptional processing. The chromosome 13 breakpoint fell in an intergenic region, ∼400 kb 3′ to the transcriptional unit of LMO7, a transcription factor gene. Targeted mutations of two other LIM/PDZ genes encoding actin-associated LIM protein or ZASP/Cypher/Oracle in mice have been found to cause congenital heart and muscle pathology.25, 26 Expression of both BNC2 and LMO7 was enriched in human fetal heart and penis compared with other tissues. However, alteration of LMO7 expression is unlikely to cause the observed phenotypes: the breakpoint was 3′ of this gene, whereas long-range position effects are generally associated with breakpoints in 5′ regulatory regions.27 Also, roughly equal expression of both LMO7 alleles was observed in the proband's lymphoblasts. BNC2 also showed biallelic expression in the proband's lymphoblasts, as judged by RT-PCR and DNA sequencing. However, BNC2 has a highly complex pattern of alternative transcriptional initiation, termination and splicing, with more than 90 000 different potential isoforms.12 Because of limited informative expressed SNPs, we were unable to assay allelic expression of specifically those BNC2 isoforms containing the last two exons most likely to be affected by the translocation. The translocation is predicted to result in a tail-to-tail orientation of BNC2 exons 1–5 and LMO7, making a fusion transcript unlikely. As the expression level of BNC2 in lymphoblastoid cells is so low (>40 cycles of amplification were required to obtain a detectable product for allelic expression assays), it is not feasible to determine whether there are any aberrant transcripts from the derivative chromosomes by techniques such as RT- or vectorette PCR. Tissues from the proband carrying the balanced translocation that express BNC2 more strongly, for example, kidney, lung and prostate, were not available.
Altered BNC2 transcription could be causally related to the proband's birth defects or coincidental. We sequenced the major coding exons of BNC2 in 40 subjects with APVR plus the proband, 48 subjects with hypospadias, and 23 controls with anatomically normal penile urethras. Missense mutations were identified in all three groups. Variants predicted by SIFT to be deleterious were present in three hypospadias subjects but also one control. The accuracy of SIFT is limited; other computational prediction programs, for example, Polyphen, add little value in the absence of structural information.22 Determining the significance of these mutations will require an assay for BNC2 function, which currently does not exist. Larger numbers of subjects need to be screened, but the data indicate that BNC2 mutations do not account for a large percentage of APVR or hypospadias.
In order to corroborate a role for BNC2 disruption in the proband's birth defects, we then examined bnc2−/− and bnc2+/− mice. No cardiovascular abnormalities were seen. Penile size was normal, suggesting normal androgen signaling in males, but both male and female homozygous mice showed a high incidence of distal urethral defects, with reduced penetrance in heterozygotes. As Bnc2 is strongly expressed in the preputial lamella and mesenchymal cells surrounding the developing os penis and urethral corpus spongiosum, the protein may act on the closure of the urethral folds. Bnc2 has been shown to be required for adequate multiplication of the palatal mesenchymal cells, and absence of bnc2 causes cleft palate. Presumably Bnc2 is essential for proper growth of periurethral mesenchymal cells, as it is for the palatal mesenchyme.21
Although different assays were used, BNC2/Bnc2 expression was detected in human but not mouse fetal heart. Early events in pulmonary vein development are very similar between humans and mouse, but the adult anatomy of the mouse pulmonary veins differs from those in humans. In the mouse, the left and right pulmonary veins come together and drain into the left atrium as a single vein, whereas in humans four veins separately enter the right atrium after intussusception of the common pulmonary vein, which makes up part of the left atrium wall.28, 29 This difference may explain why no PAPVR is seen in Bcn2-mutant mice and could suggest that the mechanism of PAPVR (which is totally unknown) may involve defects in this intussusception process.
In summary, we used a novel CGH-based approach to facilitate mapping of a balanced translocation associated with PAPVR and hypospadias. The translocation disrupted BNC2, a large, complex gene of unknown function whose expression is enriched in fetal heart and penis. The phenotype of Bnc2−/− mice corroborated a role for BNC2 in urethral development. Unlike previously identified male-specific hypospadias genes, BNC2 appears to act locally in urethral development in both sexes. Further studies are needed to determine the relationship of BNC2 expression to androgen signaling in males.
References
Gallentine ML, Morey AF, Thompson Jr IM : Hypospadias. a contemporary epidemiologic assessment. Urology 2001; 57: 788–790.
Fredell L, Iselius L, Collins A et al. Complex segregation analysis of hypospadias. Hum Genet 2002; 111: 231–234.
Page LA : Inheritance of uncomplicated hypospadias. Pediatrics 1979; 63: 788–790.
Kalfa N, Sultan C, Baskin LS : Hypospadias: etiology and current research. Urol Clin North Am 2010; 37: 159–166.
Silver RI, Russell DW : 5alpha-reductase type 2 mutations are present in some boys with isolated hypospadias. J Urol 1999; 162: 1142–1145.
Allera A, Herbst MA, Griffin JE, Wilson JD, Schweikert HU, McPhaul MJ : Mutations of the androgen receptor coding sequence are infrequent in patients with isolated hypospadias. J Clin Endocrinol Metab 1995; 80: 2697–2699.
Sutherland RW, Wiener JS, Hicks JP et al. Androgen receptor gene mutations are rarely associated with isolated penile hypospadias. J Urol 1996; 156: 828–831.
Fukami M, Wada Y, Miyabayashi K et al. CXorf6 is a causative gene for hypospadias. Nat Genet 2006; 38: 1369–1371.
Fukami M, Wada Y, Okada M et al.: Mastermind-like domain-containing 1 (MAMLD1 or CXorf6) transactivates the Hes3 promoter, augments testosterone production, and contains the SF1 target sequence. J Biol Chem 2008; 283: 5525–5532.
Mortlock DP, Innis JW : Mutation of HOXA13 in hand-foot-genital syndrome. Nat Genet 1997; 15: 179–180.
Frisen L, Lagerstedt K, Tapper-Persson M, Kockum I, Nordenskjold A : A novel duplication in the HOXA13 gene in a family with atypical hand-foot-genital syndrome. J Med Genet 2003; 40: e49.
Vanhoutteghem A, Djian P : The human basonuclin 2 gene has the potential to generate nearly 90 000 mRNA isoforms encoding over 2000 different proteins. Genomics 2007; 89: 44–58.
Romano RA, Li H, Tummala R, Maul R, Sinha S : Identification of Basonuclin2, a DNA-binding zinc-finger protein expressed in germ tissues and skin keratinocytes. Genomics 2004; 83: 821–833.
Vanhoutteghem A, Djian P : Basonuclin 2: an extremely conserved homolog of the zinc finger protein basonuclin. Proc Natl Acad Sci USA 2004; 101: 3468–3473.
Tseng H, Green H : Association of basonuclin with ability of keratinocytes to multiply and with absence of terminal differentiation. J Cell Biol 1994; 126: 495–506.
Yang Z, Gallicano GI, Yu QC, Fuchs E : An unexpected localization of basonuclin in the centrosome, mitochondria, and acrosome of developing spermatids. J Cell Biol 1997; 137: 657–669.
Tian Q, Kopf GS, Brown RS, Tseng H : Function of basonuclin in increasing transcription of the ribosomal RNA genes during mouse oogenesis. Development 2001; 128: 407–416.
Iuchi S, Green H : Basonuclin, a zinc finger protein of keratinocytes and reproductive germ cells, binds to the rRNA gene promoter. Proc Natl Acad Sci USA 1999; 96: 9628–9632.
Wang J, Zhang S, Schultz RM, Tseng H : Search for basonuclin target genes. Biochem Biophys Res Commun 2006; 348: 1261–1271.
Vanhoutteghem A, Djian P : Basonuclins 1 and 2, whose genes share a common origin, are proteins with widely different properties and functions. Proc Natl Acad Sci USA 2006; 103: 12423–12428.
Vanhoutteghem A, Maciejewski-Duval A, Bouche C et al. Basonuclin 2 has a function in the multiplication of embryonic craniofacial mesenchymal cells and is orthologous to disco proteins. Proc Natl Acad Sci USA 2009; 106: 14432–14437.
Kumar P, Henikoff S, Ng PC : Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4: 1073–1081.
Higgins AW, Alkuraya FS, Bosco AF et al. Characterization of apparently balanced chromosomal rearrangements from the developmental genome anatomy project. Am J Hum Genet 2008; 82: 712–722.
Bernheim A, Toujani S, Guillaud-Bataille M et al. Intragenic breakpoints localized by array CGH in a t(2;6) familial translocation. Cytogenet Genome Res 2007; 119: 185–190.
Nakagawa N, Hoshijima M, Oyasu M, Saito N, Tanizawa K, Kuroda S : ENH, containing PDZ and LIM domains, heart/skeletal muscle-specific protein, associates with cytoskeletal proteins through the PDZ domain. Biochem Biophys Res Commun 2000; 272: 505–512.
Guy PM, Kenny DA, Gill GN : The PDZ domain of the LIM protein enigma binds to beta-tropomyosin. Mol Biol Cell 1999; 10: 1973–1984.
Kleinjan DA, van Heyningen V : Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet 2005; 76: 8–32.
Bleyl SB, Saijoh Y, Bax NA et al. Dysregulation of the PDGFRA gene causes inflow tract anomalies including TAPVR: integrating evidence from human genetics and model organisms. Hum Mol Genet 2010; 19: 1286–1301.
Webb S, Brown NA, Wessels A, Anderson RH : Development of the murine pulmonary vein and its relationship to the embryonic venous sinus. Anat Rec 1998; 250: 325–334.
den Dunnen JT, Antonarakis SE : Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat 2000; 15: 7–12.
Acknowledgements
Human studies were funded by a UT Southwestern High Risk/High Impact grant (ARZ), NIH R01 DK59164 (LAB), and Children's Medical Center at Dallas CCRAC Grant #2006-92 (LAB). Mouse studies were funded by the Association pour la Recherche sur le Cancer and the Ligue contre le Cancer (PD).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website ()
Rights and permissions
About this article
Cite this article
Bhoj, E., Ramos, P., Baker, L. et al. Human balanced translocation and mouse gene inactivation implicate Basonuclin 2 in distal urethral development. Eur J Hum Genet 19, 540–546 (2011). https://doi.org/10.1038/ejhg.2010.245
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2010.245
Keywords
This article is cited by
-
Roles for urothelium in normal and aberrant urinary tract development
Nature Reviews Urology (2020)
-
Human basonuclin 2 up-regulates a cascade set of interferon-stimulated genes with anti-cancerous properties in a lung cancer model
Cancer Cell International (2017)
-
Fine mapping analysis confirms and strengthens linkage of four chromosomal regions in familial hypospadias
European Journal of Human Genetics (2015)