Abstract
A cis-regulatory sequence also known as zone of polarizing activity (ZPA) regulatory sequence (ZRS) located in intron 5 of LMBR1 is essential for expression of sonic hedgehog (SHH) in the developing posterior limb bud mesenchyme. Even though many point mutations causing preaxial duplication defects have been reported in ZRS, the underlying regulatory mechanism is still unknown. In this study, we analyzed the effect on transcription factor binding of a novel ZRS point mutation (463T>G) in a Pakistani family with preaxial polydactyly and triphalangeal thumb. Electrophoretical mobility shift assay demonstrated a marked difference between wild-type and the mutant probe, which uniquely bound one or several transcription factors extracted from Caco-2 cells. This finding supports a model in which ectopic anterior SHH expression in the developing limb results from abnormal binding of one or more transcription factors to the mutant sequence.
Similar content being viewed by others
Introduction
Regulatory elements are important for establishing correct expression domains in time and space. The increasing pace by which such elements are being reported leads to the natural question how these elements carry out their function. Initial identification of a balanced translocation,1 and subsequent point mutations2, 3 in families with preaxial polydactyly (PPD) and triphalangeal thumb lead to the discovery of a long-range sonic hedgehog regulatory element (ZRS), which was later also found to be duplicated in PPD patients.4, 5 ZRS is located in intron 5 of LMBR1, 1 Mb 5′ of the target gene. The ZRS is necessary for proper posterior expression in the limb.6 Mouse models with ZRS point mutations showed ectopic sonic hedgehog (SHH) expression anteriorly,2, 7 but it is still unknown how the mutations impair the normal function of the regulatory element. We hypothesized that single-nucleotide mutations in this element change the binding affinity for various transcription factors and tested it for a novel mutation detected in a three-generation family of Pakistani origin by electrophoretical mobility shift assay (EMSA).
Materials and methods
Family PD1 was examined at home in the Southern Punjab region and informed written consent was obtained from all participating individuals. This study was approved by the local ethical committee of the National Institute for Biotechnology and Genetic Engineering (NIBGE), School of Biotechnology, Quaid-i-Azam University, Pakistan. Peripheral blood samples were collected and DNA was extracted using a standard protocol (phenol:chloroform). Linkage to the known candidate region at 7q36 associated with PPD and triphalangeal thumb was performed using the highly polymorphic microsatellite markers D7S550, D7S559 and D7S2423. All samples were PCR amplified using radioactive-labeled forward primer (protocol available on request) and the alleles were size separated on 8% non-denaturing polyacrylamide gel. Direct sequencing of the 1-kb region containing ZRS was performed with the following oligonucleotide primers: forward1 5′-CCTCTGGCCAGTGTTTAAATG-3′, reverse1 5′-GGGGGAAAAAGTCATCTGGT-3′; forward2 5′-GGTGGAAGCGAAGAGTTCTG-3′, reverse2 5′-CAATTGGGGGATTGTAGAGG-3′ (TAG Copenhagen, Copenhagen, Denmark), using the dideoxy chain-termination method (BigDye ver 1.1, Applied Biosystems, Foster city, CA, USA) on an ABI 3130xl sequencer. Sequencing results were analyzed using ChromasPro software (ChromasPro 2.1, Technelysium Pty Ltd, Tewantin QLD, Australia). The identified mutation and its segregation was confirmed by agarose gel electrophoresis of MseI (New England Biolabs, Ipswich, MA, USA) digested PCR fragments.
EMSA is a standard technique commonly used to detect the interaction between proteins and γ-32P-labeled nucleic acids. It is thus well suited to test our hypothesis that the transcription factor-binding potential of the mutant ZRS sequence differs from the wild type (wt). The Caco-2 cell line derives from a cancerous human epithelial colorectal adenocarcinoma. Caco-2 cells do express SHH mRNA in both differentiated and undifferentiated stages (http://gastro.imbg.ku.dk/chipchip/). The Caco2 cell line can be used to test whether the transcription factor-binding properties of the mutated sequence differs from that of the wild type.
Standard EMSA was performed using 29-mer wild-type (T) and mutant (G) γ-32P-labeled synthetic oligonucleotide probes (Figure 1) and Caco-2 cells nuclear extract as previously described.8 Labeled probes were purified using the G25-spin column (Amersham Biosciences, Sunnyvale, CA, USA). Probe/protein complexes and unbound probes were separated by electrophoresis on 5% non-denaturing polyacrylamide gel in 0.5 × TBE buffer at 220 V for 2 h at 4°C. Gels were dried after electrophoresis and exposed in a phosphorImager (Amersham Biosciences).
Results
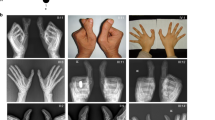
Out of 13 examined members 8 had dominantly segregating PPD and triphalangeal thumb. Four additional members were said to be affected. Confirmative radiographic examination was performed in three affected members. The metacarpals were normal in shape and number (Figure 2B). The duplicated phalanx was triphalangeal in most cases and occurred unilaterally in III-3, III-5, III-11 and III-12. The feet were normal and no other signs of dysmorphology were noticed. All individuals appeared mentally normal.
(A) Pedigree of family PD1 and haplotypes at 7q36 using microsatellite markers. The region in affected subjects segregating with the phenotype is boxed. Clinically examined individuals are shown by a horizontal line over the symbol. (B) Phenotypic appearance of affected subjects in the family PD1 with PPD triphalangial thumb. (a and b) The hands and feet of III-12, (c) clinical appearance of hands of male subject III-6, (d) radiograph of III-6 demonstrating duplicated thumbs with three phalanges.
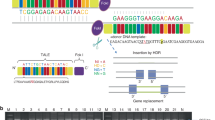
Initial STS marker analysis supported possible linkage when assuming dominant inheritance with decreased penetrance (Figure 2A). Segregation analysis of the haplotypes confirmed that all affected individuals shared the same haplotype. Two clinically normal individuals also did. Direct sequencing of ZRS showed a novel point mutation 463T>G (named according to Furniss et al,9) in intron 5 of LMBR1 (Figure 3) in all individuals sharing this haplotype. Restriction enzyme analysis confirmed the finding. The mutation was absent in 280 unrelated normal chromosomes from the same population.
(a) Representative chromatogram of the heterozygous mutation (463 T>G) in ZRS. (b) Wild type. (c) Restriction analysis using MseI restriction enzyme in all the sampled family members (normal and affected); the 268-bp mutant allele is indicated by an arrow. (d) EMSA of the conserved element demonstrating binding of Caco-2 cell line nuclear extract to both wild-type (T-allele) and mutant allele (G-allele). Unlabeled probes were added to demonstrate specific binding to both alleles. Specific binding is indicated by arrow.
EMSA analysis using Caco-2 cells nuclear extract showed specific DNA/protein binding for the mutant probes and not the wild-type probes (Figure 3d). In addition, competitive DNA/protein binding was detected using unlabeled mutant probes, whereas such competition was not detected using wild-type probe.
Discussion
PPD is one of the most frequently occurring human limb malformations. The phenotype is quite variable in affected families with a simple addition of the third phalanx leading to triphalangial thumb to whole digit duplication. Point mutations in the cis-acting regulatory element (ZRS) have been described in many families in recent years. The ZRS lies 1 Mb upstream of its target gene (SHH) and is located in intron 5 of LMBR1.2, 10 SHH is normally expressed in the zone of polarizing activity (ZPA) posteriorly in the developing limb bud. Mouse models of PPD have shown that point mutations in ZRS lead to ectopic expression of SHH at the anterior margin of limb bud.11 Heterozygous duplications including ZRS have also been described in families with triphalangial thumb-polysyndactyly syndrome.4, 5 It has been found that this long-range promoter–enhancer interaction is specific to limb bud tissues.6 It is still a matter of debate how duplications and point mutations can cause the same phenotypic defect. However, the frequency of very rare single point nucleotide changes among families and animal models with PPD clearly supports that these changes are indeed pathogenic and not only rare variants. Homozygous deletion of the ZRS element in mouse results in a complete loss of SHH expression in the limb bud leading to acheiropodia.6 Until now, 14 point mutations have been reported scattered throughout the ZRS element, suggesting that all the elements are important in regulation of SHH expression in the developing limb.2, 3, 7, 9, 10, 11 Phenotypes produced by different point mutation in ZRS are almost identical.
The nucleotide mutated in family PD1 is highly conserved between human, mouse, chicken and fugu. EMSA results for this mutation clearly demonstrate specific binding of nuclear factors to mutant probes, supporting that the pathogenic effect of the T>G alteration at position 463 acts by altering its affinity for transcription factors. Interestingly, this hypothesis was recently supported in a novel ZRS mutant mouse model (DZ), in which the authors found specific preferential binding of the transcription factor HnRNP U to the mutated sequence.12 HnRNP U is indeed expressed in Caco2 cells (http://gastro.sund.ku.dk/chipchip); however, the sequence comparison around the DZ mouse mutation (which preferentially bound to HnRNP U) and 463T>G mutation predicts that factor(s) other than HnRNP U may have bound to 463T>G mutant.12 A yeast hybrid screen using the mutant and wild-type DNA sequence as bait and based on a cDNA library derived from limb bud cells, followed by subsequent sequencing of positive clones, should be carried out to characterize the bound factor(s) in more detail.
References
Lettice LA, Horikoshi T, Heaney SJ et al: Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc Natl Acad Sci USA 2002; 99: 7548–7553.
Lettice LA, Heaney SJ, Purdie LA et al: A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet 2003; 12: 1725–1735.
Sagai T, Masuya H, Tamura M et al: Phylogenetic conservation of a limb-specific, cis-acting regulator of sonic hedgehog (Shh). Mamm Genome 2004; 15: 23–34.
Klopocki E, Ott CE, Benatar N, Ullmann R, Mundlos S, Lehmann K : A microduplication of the long range SHH limb regulator (ZRS) is associated with triphalangeal thumb-polysyndactyly syndrome. J Med Genet 2008; 45: 370–375.
Sun M, Ma F, Zeng X et al: Triphalangeal thumb-polysyndactyly syndrome and syndactyly type IV are caused by genomic duplications involving the long range, limb-specific SHH enhancer. J Med Genet 2008; 45: 589–595.
Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T : Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development 2005; 132: 797–803.
Masuya H, Sezutsu H, Sakuraba Y et al: A series of ENU-induced single-base substitutions in a long-range cis-element altering sonic hedgehog expression in the developing mouse limb bud. Genomics 2007; 89: 207–214.
Troelsen JT, Mitchelmore C, Olsen J : An enhancer activates the pig lactase phlorizin hydrolase promoter in intestinal cells. Gene 2003; 305: 101–111.
Furniss D, Lettice LA, Taylor IB et al: A variant in the sonic hedgehog regulatory sequence (ZRS) is associated with triphalangeal thumb and deregulates expression in the developing limb. Hum Mol Genet 2008; 17: 2417–2423.
Gurnett CA, Bowcock AM, Dietz FR, Morcuende JA, Murray JC, Dobbs MB : Two novel point mutations in the long-range SHH enhancer in three families with triphalangeal thumb and preaxial polydactyly. Am J Med Genet A 2007; 143: 27–32.
Lettice LA, Hill AE, Devenney PS, Hill RE : Point mutations in a distant sonic hedgehog cis-regulator generate a variable regulatory output responsible for preaxial polydactyly. Hum Mol Genet 2008; 17: 978–985.
Zhao J, Ding J, Li Y et al: HnRNP U mediates the long-range regulation of Shh expression during limb development. Hum Mol Genet 2009; 18: 3090–3097.
Acknowledgements
The Higher Education Commission of Pakistan is kindly acknowledged for funding Muhammad Farooq. The Wilhelm Johannsen Centre for Functional Genome research was established by the Danish National Research Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Farooq, M., Troelsen, J., Boyd, M. et al. Preaxial polydactyly/triphalangeal thumb is associated with changed transcription factor-binding affinity in a family with a novel point mutation in the long-range cis-regulatory element ZRS. Eur J Hum Genet 18, 733–736 (2010). https://doi.org/10.1038/ejhg.2009.225
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2009.225
Keywords
This article is cited by
-
Missing heritability of Wilson disease: a search for the uncharacterized mutations
Mammalian Genome (2023)
-
Identity-by-descent refines mapping of candidate regions for preaxial polydactyly II /III in a large Chinese pedigree
Hereditas (2018)
-
An increased duplication of ZRS region that caused more than one supernumerary digits preaxial polydactyly in a large Chinese family
Scientific Reports (2016)
-
The disruption of a novel limb cis-regulatory element of SHH is associated with autosomal dominant preaxial polydactyly-hypertrichosis
European Journal of Human Genetics (2016)
-
Biased Polyphenism in Polydactylous Cats Carrying a Single Point Mutation: The Hemingway Model for Digit Novelty
Evolutionary Biology (2014)