Abstract
The Stüve–Wiedemann Syndrome (SWS) is a frequently lethal chondrodysplasia caused by null mutations in the leukemia inhibitory factor receptor gene (LIFR) responsible for an impaired activation of the JAK–STAT pathway after LIF stimulation. Most LIFR mutations are nonsense mutations, thus prompting us to investigate the impact of aminoglycosides on the readthrough of premature termination codons (PTCs). Culturing skin fibroblasts from three SWS patients and controls for 48 h in the presence of gentamycin (200–500 μg/ml) partially restored the JAK–STAT3 pathway when stimulated by LIF. Consistently, quantitative RT-PCR analysis showed that gentamycin stabilized LIFR mRNAs carrying UGA premature termination codons. We conclude that high gentamycin concentrations can partially restore functional LIFR protein synthesis in vitro, prompting us to investigate PTC readthrough using less toxic and more efficient drugs in this presently untreatable lethal condition.
Similar content being viewed by others
Introduction
More than 1800 human inherited diseases are due to nonsense mutations leading to shortened proteins.1 To date, largely 12% of the 79 000 mutations recorded in the Human Genetic Mutation Database (HGMD) result in premature termination codons (PTCs). Several studies using various reporter systems have previously shown the ability of aminoglycosides to induce PTC readthrough and to restore a full-length protein synthesis.2, 3 These antibiotics interact with the small ribosomal RNA subunit and decrease translational accuracy, leading to a deleterious protein synthesis in prokaryotes. The same mechanism is observed for the eukaryotic translation machinery but with a lower affinity for aminoglycosides, a feature possibly accounted for by two nucleotide divergences in the small ribosomal subunit.4 When the ribosome comes across a PTC, it may substitute an amino acid to the stop codon and resume protein synthesis.
Several in vivo studies using aminoglycosides have successfully induced PTC readthrough in genetic disorders including Duchenne muscular dystrophy and cystic fibrosis.5, 6, 7 Moreover, preliminary in vivo studies using parenteral gentamycin were associated with clinical benefits and a significant improvement of CFTR-mediated chloride transport function in nasal and sweat gland epithelia.8 Recessive genetic disorders caused by nonsense mutations are good candidates for aminoglycoside readthrough, as small amounts of functionally active protein may have a clinical impact.7
We have previously identified the leukemia inhibitory factor receptor (LIFR) as the gene responsible for the Stüve–Wiedemann Syndrome (SWS (MIM 601559)) and have shown that LIFR mutations are responsible for an impairment of JAK–STAT signalling through LIF induction.9 This autosomal recessive disorder belongs to the group of bent-bone dysplasias and is characterized by a bowing of the lower limbs with an internal cortical thickening, wide metaphyses with an abnormal trabecular shape and camptodactyly.10 Other clinical features include respiratory distress, feeding and swallowing difficulties, and hyperthermic episodes responsible for death in the first months of life. This condition results from a loss of functional mechanism with a fair number of premature stop mutations. The LIF receptor is a heterodimer composed of the gp190 subunit (also named LIFR) and the gp130 subunit. Binding of LIF to the LIFR complex induces signalling through the JAK–STAT and MAPK pathways (Figure 1a). Upon phosphorylation on the tyrosine 705 residue by JAK, STAT3 molecules dimerize and translocate into the nucleus, where they bind to cytokine-responsive elements and ultimately activate gene transcription.
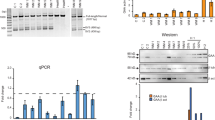
(a) JAK–STAT3 pathway activation by LIF. (b) Partial leukemia inhibitory factor receptor (LIFR) sequences of three patients with the Stüve-Wiedemann Syndrome (SWS). Numbers represent nucleotide positions in LIFR cDNA. Premature termination codons are boxed and footnoted with an asterix. The fourth nucleotide just downstream of premature termination codon (PTC) is shaded. Amino-acid sequences are in italics and mutation is indicated by a vertical arrow. Sequence consensus was determined in Saccharomyces Cerevisiae12 and promotes> 5% readthrough efficiency. (c) Western-blot analysis using anti-phospho-STAT3 and anti-actin antibodies in patient fibroblasts with nonsense mutations and control fibroblasts.
Here, we show that gentamycin induced PTC readthrough of LIFR mRNA in cultured skin fibroblasts of SWS patients.
Materials and methods
Fibroblast cultures
Three SWS patients carrying homozygous nonsense mutations displayed an impaired JAK–STAT signalling after LIF stimulation in cultured skin fibroblasts.9 Premature termination codons were located at either codon 597 (patients 1 and 2, c.1789>T)) or codon 812 of the LIFR mRNA (patient 3, c.2434>T, Figure 1b). The premature termination codons were of the TGA type in all three cases. The fourth nucleotide located just downstream of the PTC was a cytosine (the most efficiently passed stop codon, patients 1 and 2) or a guanine (patient 3).
Skin biopsies were obtained after obtaining informed consent and grown until confluence in RPMI 1640 (Invitrogen, Carlsbad, CA, USA) added with 10% fetal bovine serum. Aminoglycosides (200 and 500 μg/ml gentamycin) were added during 48 h. The cells were then rinsed with phosphate buffered saline (PBS) and incubated for 12 h in a serum-free medium (with gentamycin) to remove LIFR-binding cytokines. Fibroblasts were washed with PBS, incubated in serum-free medium for 15 min with 20 ng/ml LIF, washed twice with PBS and harvested. Western blot analyses of fibroblast lysates were carried out using an antiphospho (TYR705)-STAT3 antibody.
Western blots
Cells were lysed for 1 h at 4°C with a radioimmunoprecipitation buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P40, 0.5% sodium deoxycholate, 2 mM sodium orthovanadate and protease inhibitor cocktail). Proteins were quantified using the Bradford method. A total of 30 μg of proteins were loaded onto a 4–12% gradient polyacrylamide gel (Invitrogen), blotted on a PVDF membrane and incubated with antiphospho-STAT3 and anti-actin antibodies (Tebu, Le Perray en Yvelines, France). Western blots were revealed with the ECL kit as recommended by the manufacturer (GE Healthcare, Little Chalfont, UK).
Quantitative RT-PCR
Fibroblasts were grown until confluence and treated with gentamycin (200 μg/ml) for 48 h or with cycloheximide (200 μg/ml) for 5 h. Total RNAs were extracted with Trizol as recommended by the manufacturer (Invitrogen). RNA was quantified with a Nanodrop apparatus and its quality was analyzed on an agarose gel. To amplify the LIFR mRNA, RT-PCR was carried out using the following primers: forward 5′-TTCTGGTCTCGAAGAGTGGAGT-3′ and reverse 5′-CAGTTTGTATGGCCAATCAGTG-3′ (product length of 169 bp). Beta-actin was also amplified using the following primers: forward 5′-AGATCAAGATCATTGCTCCTCCTG-3′; reverse 5′-AAAACAAATAAAGCCATGCCAATCT-3′ (product length of 243 bp). We performed real-time PCR, using LightCycler technology (Roche Manheim, Manheim, Germany). Experiments were carried out in triplicate with the total mRNA from two fibroblast cultures. Student's t-test was used for the determination of P values.
Results
Figure 1c shows that STAT3 failed to be phosphorylated in response to LIF in cultured SWS fibroblasts. By contrast, the addition of either 200 μg/ml or 500 μg/ml of gentamycin to the medium for 48 h partially restored STAT3 phosphorylation on the tyrosine 705 residue, consistent with the synthesis of trace amounts of functional LIFR protein. This result indicated that gentamycin was able to induce a readthrough of the UGA premature termination codon of LIFR mRNA. Cultured fibroblasts from patients 2 and 3 incubated with a high gentamycin concentration (500 μg/ml) showed a weaker signal than those incubated with lower concentrations of gentamycin (200 μg/ml). Consistently, a microscopic observation of the cells cultured in the presence of 500 μg/ml of gentamycin showed abnormal round-shaped fibroblasts, suggestive of a toxic effect of gentamycin (data not shown).
As a nonsense mediated decay (NMD) mechanism could also account at least in part for a reduction of the specific protein, we quantified LIFR mRNA in SWS and control fibroblasts using quantitative PCR. A reverse transcriptase PCR analysis of LIFR transcripts demonstrated a weak signal in patients 1–3 compared with control fibroblasts (data not shown). We subsequently performed real time PCR experiments in three patients and in one control in duplicate. The amounts of LIFR mRNA were normalized for the amount of β-actin mRNA. The stability of LIFR mRNA was decreased to 4–16% of control values in the three patients (P<0.005) (Figure 2). Consistently, an incubation of patient fibroblasts with 200 μg/ml cycloheximide, a known NMD inhibitor,11 restored 70–200% of the LIFR mRNA amount compared with that in controls. Incubating patient fibroblasts with 200 μg/ml gentamycin also restored a significant amount of LIFR mRNA (Figure 2).
Quantitative analysis of leukemia inhibitory factor receptor (LIFR) gene expression in fibroblasts from control and patients 1–3. The relative amounts of LIFR mRNA levels compared with β-actin mRNA levels were obtained for controls and for the three patients untreated and treated with gentamycin (200 μg/ml) or cycloheximide (200 μg/ml). The results shown were means±SDs from two independent experiments. In each experiment, samples were run in triplicate.
Discussion
Studying the stimulation of the STAT3 pathway by LIF in cultured skin fibroblasts of three SWS patients, we have shown that gentamycin was able to partially restore functional LIFR synthesis. Although we did not demonstrate by western blot the presence of the full-length LIFR protein, the stimulation of the STAT3 pathway was substantial, allowing us to conclude a functional LIFR synthesis, as LIF cytokine activates the STAT3 pathway only through LIFR. All patients tested carried a TGA-type stop codon, which is reported as the PTC most efficiently bypassed by ribosomes. In addition, in two of three patients, the fourth nucleotide downstream of the stop codon was a cytosine, which is the most readily bypassed nucleotide.12, 1 We obtained better results using lower gentamycin concentrations in two of three patients, illustrating the toxic effect of high doses of gentamycin on cultured fibroblasts. Unfortunately, such doses of gentamycin (200 or 500 μg/ml) cannot be achieved in human serum in vivo, as this drug causes ototoxicity at serum concentrations above 12 μg/ml.13 Indeed gentamycin seems to generate free radicals within the inner ear, with a subsequent permanent damage to sensory cells and neurons, resulting in a permanent hearing loss.14 Gentamycin also induces nephrotoxicity, which is characterized by slow rises in serum creatinine, tubular necrosis and marked decreases in the glomerular filtration rate.15
A lower gentamycin concentration could be nevertheless beneficial for patients,16 although it was not possible to detect the full-length protein by western blot analysis. This approach may be interesting in recessive disorders when trace amounts of functional proteins are expected to have in vivo effects.
We have also demonstrated that LIFR mRNA of SWS patients was subjected to NMD, seriously altering the specific mRNA level. Our study also supports the view that gentamycin inhibited NMD and therefore increased the amount of PTC containing LIFR mRNA, which could be therefore submitted to a readthrough. This inhibition of NMD by gentamycin has been previously suggested,17 and recently, Allamand et al18 reported the significant stabilization of laminin α2 chain mRNA by negamycin, a dipeptide antibiotic also interacting with ribosome. Finally, we carried out our studies in skin fibroblasts and we cannot exclude the fact that NMD efficiency is different in cartilage as NMD can vary from one tissue to the other.
The toxicity of gentamycin prompts a search for less-toxic drugs such as negamycin, which may also interact with ribosomes and promote a readthrough. A compound named PTC124 has been developed by PTC Therapeutics, South Plainfield, NJ, USA.19 This drug has been successfully used in mouse models of cystic fibrosis and in cultured cells from muscle biopsies of patients affected by Duchenne muscular dystrophy.20 This study showed an equal inducing effect of the molecule on readthrough compared with aminoglycosides associated with reduced toxicity and a better oral bioavailability. Yet, this molecule does not seem to inhibit the NMD mechanism.
We conclude that gentamycin is able to restore trace amounts of functional LIFR synthesis by enhancing PTC readthrough and inhibiting NMD. Ongoing studies will hopefully lead to the identification of less toxic drugs with similar therapeutic effects.
Conflict of interest
The authors declare no conflict of interest.
References
Zingman LV, Park S, Olson TM, Alekseev AE, Terzic A : Aminoglycoside-induced translational read-through in disease: overcoming nonsense mutations by pharmacogenetic therapy. Clin Pharmacol Ther 2007; 81: 99–103.
Manuvakhova M, Keeling K, Bedwell DM : Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA 2000; 6: 1044–1055.
Lai CH, Chun HH, Nahas SA et al: Correction of ATM gene function by aminoglycoside-induced read-through of premature termination codons. Proc Natl Acad Sci USA 2004; 101: 15676–15681.
Fan-Minogue H, Bedwell DM : Eukaryotic ribosomal RNA determinants of aminoglycoside resistance and their role in translational fidelity. RNA 2008; 14: 148–157.
Barton-Davis ER, Cordier L, Shoturma DI, Leland SE, Sweeney HL : Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J Clin Invest 1999; 104: 375–381.
Politano L, Nigro G, Nigro V et al: Gentamicin administration in Duchenne patients with premature stop codon. Preliminary results. Acta Myol 2003; 22: 15–21.
Wilschanski M, Yahav Y, Yaacov Y et al: Gentamicin-induced correction of CFTR function in patients with cystic fibrosis and CFTR stop mutations. N Engl J Med 2003; 349: 1433–1441.
Sermet-Gaudelus I, Renouil M, Fajac A et al: In vitro prediction of stop-codon suppression by intravenous gentamicin in patients with cystic fibrosis: a pilot study. BMC Med 2007; 29: 5.
Dagoneau N, Scheffer D, Huber C et al: Null leukemia inhibitory factor receptor (LIFR) mutations in Stüve-Wiedemann/Schwartz-Jampel type 2 syndrome. Am J Hum Genet 2004; 74: 298–305.
Wiedemann HR, Stüve A : Stüve-Wiedemann syndrome: update and historical footnote. Am J Med Genet 1996; 63: 12–16.
Mitchell P, Tollervey D : An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′ → 5′degradation. Mol Cell 2003; 5: 1405–1413.
Namy O, Hatin I, Rousset JP : Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep 2001; 2: 787–793.
Begg EJ, Barclay ML : Aminoglycosides-50 years on. Br J Clin Pharmacol 1995; 39: 597–603.
Selimoglu E : Aminoglycoside-induced ototoxicity. Curr Pharm Des 2007; 13: 119–126.
Martinez-Salgado C, Lopez-Hernandez FJ, Lopez-Novoa JM : Glomerular nephrotoxicity of aminoglycosides. Toxicol Appl Pharmacol 2007; 223: 86–98.
Kellermayer R : Translational readthrough induction of pathogenic nonsense mutations. Eur J Med Genet 2006; 49: 445–450.
Kaufman RJ : Correction of genetic disease by making sense from nonsense. J Clin Invest 1999; 104: 367–368.
Allamand V, Bidou L, Arakawa M et al: Drug-induced readthrough of premature stop codons leads to the stabilization of laminin alpha2 chain mRNA in CMD myotubes. J Gene Med 2008; 10: 217–224.
Welch EM, Barton ER, Zhuo J et al: PTC124 targets genetic disorders caused by nonsense mutations. Nature 2007; 447: 87–91.
Du M, Liu X, Welch EM, Hirawat S, Peltz SW, Bedwell DM : PTC124 is an orally bioavailable compound that promotes suppression of the human CFTR-G542X nonsense allele in a CF mouse model. Proc Natl Acad Sci 2008; 105: 2064–2069.
Acknowledgements
This work was supported by a grant (N 12811) from ‘Association Française contre les Myopathies’ (AFM). Samuel Bellais is the recipient of a grant from the AFM (N 12807).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bellais, S., Le Goff, C., Dagoneau, N. et al. In vitro readthrough of termination codons by gentamycin in the Stüve–Wiedemann Syndrome. Eur J Hum Genet 18, 130–132 (2010). https://doi.org/10.1038/ejhg.2009.122
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2009.122
Keywords
This article is cited by
-
Advances in therapeutic use of a drug-stimulated translational readthrough of premature termination codons
Molecular Medicine (2018)
-
Stüve-Wiedemann syndrome: LIFR and associated cytokines in clinical course and etiology
Orphanet Journal of Rare Diseases (2014)