Abstract
To date, six genes are known to cause nemaline (rod) myopathy (NM), a rare congenital neuromuscular disorder. In an attempt to find a seventh gene, we performed linkage and subsequent sequence analyses in 12 Turkish families with recessive NM. We found homozygosity in two of the families at 1q12-21.2, a region encompassing the γ-tropomyosin gene (TPM3) encoding slow skeletal muscle α-tropomyosin, a known NM gene. Sequencing revealed homozygous deletion of the first nucleotide of the last exon, c.913delA of TPM3 in both families. The mutation removes the last nucleotide before the stop codon, causing a frameshift and readthrough across the termination signal. The encoded αTmslow protein is predicted to be 73 amino acids longer than normal, and the extension to the protein is hypothesised to be unable to form a coiled coil. The resulting tropomyosin protein may therefore be non-functional. The affected children in both families were homozygous for the mutation, while the healthy parents were mutation carriers. Both of the patients in Family 1 had the severe form of NM, and also an unusual chest deformity. The affected children in Family 2 had the intermediate form of NM. Muscle biopsies showed type 1 (slow) fibres to be markedly smaller than type 2 (fast) fibres. Previously, there had been five reports, only, of NM caused by mutations in TPM3. The mutation reported here is the first deletion to be identified in TPM3, and it is likely to be a founder mutation in the Turkish population.
Similar content being viewed by others
Introduction
The congenital myopathies include a spectrum of neuromuscular disorders defined on the basis of structural abnormalities in the muscle fibres. The most common of these disorders is the genetically and clinically heterogeneous nemaline (rod) myopathy (NM). The main symptom is muscle weakness, and muscle biopsy shows disorganisation of the muscle Z discs and accumulation of nemaline bodies, that is, aggregates consisting of filament proteins originating from sarcomeric Z discs.1 NM has been divided into six clinical categories varying from very severe, sometimes even lethal forms, to milder muscle disease (MIM 609284, 256030, 161800, 609285, 605355, and 609273).2, 3
The six known NM genes all encode proteins for the thin filament of the muscle sarcomere: nebulin (NEB, MIM 161650),4 α-skeletal muscle actin (ACTA1, MIM 102610),5 α-slow tropomyosin (TPM3, MIM 191030),6 β-tropomyosin (TPM2, MIM 190990),7 slow troponin T (TNNT1, MIM 191041),8 and cofilin 2 (CFL2, MIM 601443).9 Mutations in NEB and ACTA1 cause most of the NM cases. Mutations in TNNT1, TPM2, TPM3 and CFL2 are rare. While mutations in tropomyosins have been reported in a few families each, TNNT1 and CLF2 mutations have been found respectively only in the Amish and in a singleton family.
In an attempt to find a seventh gene for NM, we performed a linkage study using samples from 12 Turkish families with consanguinity or familial NM. Our hypothesis was that there could be a Turkish founder mutation in either a known or a novel NM gene. Two of the families were found to be homozygous for the same haplotype around the TPM3 locus on chromosome 1, and subsequent sequencing of TPM3 identified the same deletion in both of the families.
Materials and methods
Subjects
Family 1
In Family 1, the healthy parents were not known to be related but originated from the same small village. There were three children, two affected boys with severe NM and one younger, healthy boy. Both affected boys were born with contractures of the knees and ankles and showed delayed motor development. Specifically, weakness of the neck muscles was noted. The older brother (17 years) did not achieve walking, while the younger brother (12 years) walked slowly with a waddling gait from the age of 2.5–3 years. There was no evidence of cardiac involvement. Restricted vital capacity was documented in both patients who received nocturnal non-invasive ventilation from the beginning of the second decade. They had a peculiar pectus carinatum deformity and scoliosis, and facial weakness (Figure 1a). A muscle biopsy of the older brother showed hypotrophic type 1 fibres containing nemaline bodies, a nerve biopsy did not display significant pathological changes.
(a) The younger of the two affected brothers of family 1 at age 11. The severe, unusual chest deformity (pectus carinatum) can be seen. There is pronounced weakness of neck muscles and also facial weakness. (b) The affected children of family 2 have pronounced facial weakness and foot drop. The children are 5 and 6 years of age at the time that the picture was taken.
Family 2
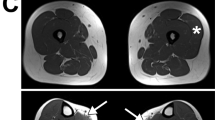
The parents of the two affected children of Family 2 are second cousins. Both children had muscle hypotonia during the first month of life. Particular features were pronounced facial weakness, lack of head control, lax distal joints, and scoliosis already visible during the first year (Figure 1b). Motor milestones were delayed; at 2 years of age, the children were able to sit unaided, but unable to reach the sitting position independently. The boy achieved his best motor function, taking a few steps with support, between four and 6 years of age. The girl was a little better and walked unaided for short distances from age 2–3 years to 6–7 years, but even at this stage she was unable to get up to a sitting position independently. At the age of 6 years, both children could lift their hands to their mouths, but could not elevate their arms completely. At the age of 10 (the girl) and 11 (the boy), they were unable to feed themselves and needed help for dressing and undressing. The joints were hyperextendable, and from the age of 8 years onwards, both children developed contractures, first of the ankles, then of the knees and elbows. Both have developed a mild chest deformity over time. At age 13 (the boy) and 12 years (the girl), they were started on non-invasive ventilation; vital capacity was 28%. The girl had suffered three pneumonias in the years before. At age 13 and 14, thoracic surgery was considered but not performed as vital capacity was <20% and the procedure was considered too dangerous. The diagnosis of NM was established by muscle biopsy in the girl when she was aged 3 months and admitted to hospital for bronchitis. The biopsy showed the type 1 fibres to be markedly smaller than the type 2 fibres (Figure 2a). The biopsy slides were no longer available for precise calculation of the fibre size disproportion. Electron microscopy showed nemaline bodies in the hypotrophic fibres (Figure 2b). Her older brother at this time was aged 15 months and was unable to walk. The children were equipped with electric wheelchairs at the age of 5 (the girl) and 6 years (the boy).
(a) Light microscopic image of the NADH–TR-stained muscle of the proband of family 2 showing disproportion of fibre size with darkly stained type 1 fibres (arrow) being smaller than the more lightly stained type 2 fibres (arrow head). (b) Numerous nemaline bodies (arrow) in hypotrophic muscle fibres in the electron microscope images.
DNA samples were available from all members of both families. After the identification of the mutation, we screened 16 further probands of Turkish NM families, and 120 control individuals for the mutation.
Linkage studies
To exclude from the search for a seventh NM gene in those Turkish families that might have a mutation in a known NM gene, we analysed the families for linkage to the known NM gene loci, and also to the mixed core-rod myopathy ryanodine receptor (RYR1) (OMIM 180901) locus on chromosome 19,10, 11 and to the core-rod locus on chromosome 15.12
In the chromosomal region 1q12-21.2 where TPM3 is located, we analysed eight microsatellite markers encompassing a region of 18 cM; D1S252, D1S498, D1S2347, D1S2858, D1S305, D1S2624, D1S2635, and D1S484. In addition to TPM3, other muscle genes located within this region include the transformation suppressor gene (YL-1, MIM 600607), and the tropomodulin 4 (muscle) gene (TMOD4, MIM 605834).
The subsequent search for the seventh NM gene was performed as a genome-wide linkage analysis using 358 microsatellite markers covering all autosomes. The distance between these markers was on average 10 cM. This work was carried out at the Finnish Genome Centre.
Mutation analyses
PCR
To perform mutation screening for TPM3 by sequencing, we designed primers to amplify in PCR reactions the exons expressed in the muscle isoform of the protein (TPM3 sequence sources in GenBank: cDNA accession no. X04201, protein accession no. P05753, and accession no. NM_152263). We used AmpliTaq Gold as the polymerase enzyme for the PCR reactions. Detailed PCR reaction conditions will be provided on request.
RNA extraction from muscle biopsy and RT-PCR
To verify the presence of the alteration in the mRNA of the muscle isoform of TPM3, we performed RT-PCR using a frozen muscle biopsy sample from the proband of Family 2. Total RNA was extracted from the muscle biopsy using the RNeasy Mini kit (Qiagen Sciences, MD, USA) including DNase treatment. To sequence the whole length of the TPM3 cDNA expressed in the patient, we used the primers designed and published by Wattanasirichaigoon et al.13 We designed additional primers from exon 7 to exon 9b in the 3′UTR region to verify the presence of the 1 bp deletion in the mRNA as well. Primer information will be provided on request.
Sequencing
The purified PCR products were sequenced using BigDye version 3.1 sequencing chemistry and an ABI 3730 DNA Analyzer (Applied Biosystems, Foster City, USA). The sequences were analysed using Sequencher 4.5 software.
Coiled-coil analysis using the COILS tool
To perform in silico prediction of the effect of the c.913delA mutation on the αTmslow protein coiled-coil structure, we used the tool COILS, version 2.2. The effect of the c.915A>C mutation published by Wattanasirichaigoon et al13 was also analysed using the COILS program for comparison.14, 15, 16 The COILS tool, version 2.2, is available at www.ch.embnet.org/software/coils/COILS_doc.html.
Results
Linkage results
Linkage analysis showed homozygosity in the TPM3 region on chromosome 1 in two of the families. Haplotypes and linkage results in the chromosomal region 1q12-21.2 are shown together with the pedigrees in Figure 3. The two families shared the same homozygous haplotype for the markers D1S498 and D1S305. The distance between these two markers is 2.4 cM. This is the minimum length of the shared haplotype, while the maximum shared haplotype, the distance between the markers D1S252 and D1S2624 for which the patients were no longer homozygous, is 11.28 cM.
There was no other shared haplotype at any of the remaining known NM gene loci. However, only the NEB locus could be excluded with certainty in two families. Genome-wide screening in the remaining cohort also did not show any significant shared haplotype for any two or more families anywhere else in the genome.
Sequencing results
Sequence analysis of TPM3 in the affected sib pairs revealed a homozygous deletion of the first nucleotide, an adenine, of the last exon (exon 9b) of the TPM3 isoform expressed in skeletal muscle, c. 913delA (Figure 4a and b)6, 13, 17, 18, 19. All four parents were found to be healthy mutation carriers.
(a) Schematic illustration of the TPM3 gene and mutations identified in it to date. The boxes represent exons; black boxes represent exons expressed in all TPM3 isoforms, grey represent the muscle-specific exons, and white boxes represent exons not present in the muscle TPM3 isoform αTmslow. The lines show the splicing pattern of TPM3 for the isoform αTmslow. Letters mark the NM-causing mutations identified in TPM3. A=heterozygous missense mutation p.M8R,6, 18 B=homozygous mutation p.Q31X,17 C=heterozygous p.R167H, D=compound heterozygous splice site mutation in intron 9a c.855G>A together with E=another compound heterozygous mutation; p.X285NextX57 identified in one patient.13 F=the mutation described in this article – a homozygous frameshift mutation eliminating the stop codon p.X285NextX74. Protein accession no. P05753, cDNA accession no. X04201; exon numbering as in Dufour et al.19 (b) The sequence chromatogram of the intron 9a – exon 9b boundary of TPM3. The WT (topmost), the sequence of a heterozygous carrier (on line two ; ‘A’ underlined is deleted from one of the alleles), the sequence of a homozygous patient (on line three).
RT-PCR from muscle tissue confirmed the presence of the mutation in the cDNA. The deletion of the last nucleotide before the stop codon disrupts the reading frame, causing readthrough across the position of the normal stop codon, which is predicted to result in an αTmslow elongated by 73 amino acids (p.X285NextX74).
Sequencing samples from probands of the 10 remaining Turkish families in our series, and 6 from additional Turkish families, making a total of 18 families, the same deletion was not found neither in any other families nor in the 120 control individuals studied.
In the patients included in this linkage-based study, no mutations were found by sequencing of other known NM genes. ACTA1 and all the exons of TPM3 were sequenced in all the families. Mutation analysis of NEB has been completed in two of the families, and no mutations were found.
The results of the COILS analysis
The 73 amino-acid C-terminal elongation (NYHRFCSVLDLPPLLLGEPKAPLSLWIPFGSAWLVPKALGWGSKKQLMYFLPPPPQIKMLSCWKPHATLHLCH) of the αTmslow protein is predicted by the COILS program not to be able to form a coiled-coil structure. We obtained the same prediction for the previously published mutation c.915A>C,13 which elongates the C-terminus of αTmslow with 57 amino acids (SLSPFLLCSGSAPFTPRGTQGPTLALDSIWVSLAGPQGIRMGEQKATYVFSST PTPN).
Discussion
The mutation identified in this study is the first deletion to be identified in TPM3, and also the first homozygous deletion to be identified in any of the tropomyosin genes. This mutation is likely to be a founder mutation in the Turkish population.
The tropomyosins exist as double-stranded coiled-coil homo- or heterodimers forming head-to-tail polymers, along the actin filament. The main roles of tropomyosins in muscle are to stabilise the sarcomeric thin (actin) filament20 and to regulate muscle contraction.21 The tropomyosin protein family of more than 40 different tropomyosin isoforms is encoded by four different genes; αTm, that is, TPM1 (MIM 191010), βTm, that is, TPM2, γTm, that is, TPM3, and δTm, that is, TPM4 (MIM 600317),22 which have a potential to generate all these isoforms due to the use of different promoters or variable intragenic splicing.22, 23, 24 TPM1, TPM2, and TPM3 encoding the isoforms αTmfast, βTm, and αTmslow are expressed in skeletal muscle. TPM2 is expressed in both slow (type 1), and, to a lesser extent, fast (type 2) muscle fibres, whereas TPM3 encodes the slow-specific isoform αTmslow.25
Mutations in slow muscle fibre-specific TPM3 cause NM6, 13, 17, 18, 26 and have recently been shown to perhaps be a relatively common cause of congenital fibre type disproportion (CFTD),27 while mutations of TPM2, expressed in all muscle fibres, have been reported in patients either with NM,7 distal arthrogryposis,28 or cap disease.29 TPM1 mutations are known to cause dilated and hypertrophic cardiomyopathies (MIM 115196).30, 31, 32
The five previously published NM mutations in TPM3 include both recessively (AR) and dominantly (AD) inherited forms of NM, among these the first NM-causing mutation identified by Laing et al.6 This was the mutation p.M8R in a large family with a juvenile onset, AD form of NM predominantly affecting the lower limbs. Another AD NM mutation, p.R167H, has been reported first as a de novo mutation in a sporadic case, again with a predilection of weakness for the lower limbs,26 and the same mutation was recently identified in a large French family.18 The French patients had typical or mild forms of NM. In addition, two patients with AR NM have been described; the first had a homozygous nonsense mutation p.Q31X and another two different compound heterozygous mutations (c.855-1G>A (splice site mutation) and p.X285S). The former patient had a severe and the latter had the intermediate form of NM.13, 17 (Figure 4a).
The mutation described here causes a frameshift and eliminates the normal stop codon of TPM3. A different mutation altering the stop codon has been published previously. That mutation is predicted to make the tropomyosin 57 amino acids longer and was confirmed by western blotting to alter the size of the protein produced.13 This patient was a compound heterozygote and the second mutation was a splice site mutation in the exon 9sk splice acceptor site leading to skipping of the last exon of the muscle isoform of TPM3 and inclusion of the last exon of the non-muscle specific isoform of TPM3. Unfortunately, muscle biopsies were not available from any of our patients for protein analysis, but RT-PCR and sequencing showed the mutation to be present at the mRNA level. Therefore, it appears likely that an elongated αTmslow protein is expressed in our patients as well.
The present mutation is expected to render the tropomyosin protein non-functional. Seventy-three amino acids are added to a protein normally 285 amino acids long, an approximately 25% increase in protein length. The amino-acid sequence translated from the 3′UTR due to the frameshift is not predicted to be able to form a coiled-coil and the C-terminal sequence of the tropomyosin protein is altered. The C-terminal and N-terminal sequences of tropomyosin are crucial to the normal head-to-tail polymerisation of tropomyosin.33
Therefore, it might be surmised that the tropomyosin resulting from the mutation described here, with added uncoiled protein at the C-terminal end of the molecule, would not be able to form the coiled-coil polymer that normally runs the length of the thin filament. On the other hand, tropomyosins fused to non-coiled-coil fluorescent proteins can incorporate into sarcomeres suggesting that the mutant tropomyosin in these families might not be completely non-functional.34 If, however, the extended mutant α-tropomyosin in the present patients is non-functional, it would make these patients comparable to the NM patient homozygous for an early nonsense mutation in the TPM3 gene described by Tan et al.17 The present patients would then have no functional slow α-tropomyosin in their slow, type 1, muscle fibres, explaining why the type 1 muscle fibres are severely hypotrophic. The only functional tropomyosin in the patients' type 1 fibres would be whatever β-tropomyosin is present. Fast, type 2 muscle fibres do not express TPM3, and are therefore relatively unaffected, as seen in the patients' muscle biopsies. Although there is a wide variability in histology and clinical features in patients with TPM3 mutations, a similarity is that the patients reported hitherto consistently show smallness and definite abnormality of type 1 fibres. The mutation in the present patients should also affect the function of the cytoskeletal isoforms Tm5NM3 and Tm5NM11, which include the mutated exon and this too may play a role in the overall clinical phenotype.25
On the basis of clinical and histological data of previously reported cases and of the two families described here, it appears that dominant mutations of TPM3 give rise to milder forms of NM than recessively inherited mutations. The phenotype as well as the histology does, however, vary between the AR and AD NM cases and even between patients with exactly the same mutation. The dominant mutation p.R167H reported first as a de novo mutation in one patient and recently segregating in a large French family caused different histological patterns. The affected children in family 1 in this article have an unusual chest deformity, while in the patients of family 2, the chest deformity is only mild. Still, both of the families have the same homozygous deletion eliminating the stop codon of the muscle-specific TPM3. The underlying reason for this clinical variability in patients with the same mutation remains unclear. A significant component of the variability may result from diffrences in splicing of alternative exons between different individuals as creation of different isoforms through alternative splicing is extensive in the tropomyosins.25 Variation in expression of the different tropomyosin genes in different individuals, as well as the effects of other muscle genes, modifier genes, and/or normal variability in the genome may all influence the patients' outcome.
Our results of linkage studies in AR NM (unpublished observations) and studies by others in AD NM,35 though the latter did not specifically exclude the then unknown CFL2, indicate that there is probably a seventh NM gene yet to discover.
Both of the families described here with the 1 bp deletion in TPM3 are Turkish families residing in Germany. They are not known to be related, and they originate from villages located some 300 km apart, but the shared haplotype indicates that the mutation discussed is a founder mutation. We did not, however, detect this mutation in any of the additional 16 Turkish families studied. These families also showed no other mutation in TPM3. Thus, we succeeded in identifying a founder mutation in the Turkish families, but only in a known NM gene, and only accounting for the NM in some Turkish families. Nevertheless, we suggest that it is warranted to examine NM patients of Turkish origin for this mutation.
References
Shy GM, Engel WK, Somers JE, Wanko T : Nemaline myopathy: a new congenital myopathy. Brain 1963; 86: 793–810.
Wallgren-Pettersson C, Pelin K, Hilpela P et al: Clinical and genetic heterogeneity in autosomal recessive nemaline myopathy. Neuromuscul Disord 1999; 9: 564–572.
Wallgren-Pettersson C, Laing NG : Report of the 70th ENMC International Workshop: nemaline myopathy, 11–13 June 1999, Naarden, The Netherlands. Neuromuscul Disord 2000; 10: 299–306.
Pelin K, Hilpela P, Donner K et al: Mutations in the nebulin gene associated with autosomal recessive nemaline myopathy. Proc Natl Acad Sci USA 1999; 96: 2305–2310.
Nowak KJ, Wattanasirichaigoon D, Goebel HH et al: Mutations in the skeletal muscle alpha-actin gene in patients with actin myopathy and nemaline myopathy. Nat Genet 1999; 23: 208–212.
Laing NG, Wilton SD, Akkari PA et al: A mutation in the alpha tropomyosin gene TPM3 associated with autosomal dominant nemaline myopathy. Nat Genet 1995; 9: 75–79.
Donner K, Ollikainen M, Ridanpaa M et al: Mutations in the beta-tropomyosin (TPM2) gene–a rare cause of nemaline myopathy. Neuromuscul Disord 2002; 12: 151–158.
Johnston JJ, Kelley RI, Crawford TO et al: A novel nemaline myopathy in the Amish caused by a mutation in troponin T1. Am J Hum Genet 2000; 67: 814–821.
Agrawal PB, Greenleaf RS, Tomczak KK et al: Nemaline myopathy with minicores caused by mutation of the CFL2 gene encoding the skeletal muscle actin-binding protein, cofilin-2. Am J Hum Genet 2007; 80: 162–167.
Monnier N, Romero NB, Lerale J et al: An autosomal dominant congenital myopathy with cores and rods is associated with a neomutation in the RYR1 gene encoding the skeletal muscle ryanodine receptor. Hum Mol Genet 2000; 9: 2599–2608.
Scacheri PC, Hoffman EP, Fratkin JD et al: A novel ryanodine receptor gene mutation causing both cores and rods in congenital myopathy. Neurology 2000; 55: 1689–1696.
Gommans IM, Davis M, Saar K et al: A locus on chromosome 15q for a dominantly inherited nemaline myopathy with core-like lesions. Brain 2003; 126: 1545–1551.
Wattanasirichaigoon D, Swoboda KJ, Takada F et al: Mutations of the slow muscle alpha-tropomyosin gene, TPM3, are a rare cause of nemaline myopathy. Neurology 2002; 59: 613–617.
Parry DA : Coiled-coils in alpha-helix-containing proteins: analysis of the residue types within the heptad repeat and the use of these data in the prediction of coiled-coils in other proteins. Biosci Rep 1982; 2: 1017–1024.
Lupas A, Van Dyke M, Stock J : Predicting coiled coils from protein sequences. Science 1991; 252: 1162–1164.
Lupas A : Coiled coils: new structures and new functions. Trends Biochem Sci 1996; 21: 375–382.
Tan P, Briner J, Boltshauser E et al: Homozygosity for a nonsense mutation in the alpha-tropomyosin slow gene TPM3 in a patient with severe infantile nemaline myopathy. Neuromuscul Disord 1999; 9: 573–579.
Penisson-Besnier I, Monnier N, Toutain A, Dubas F, Laing N : A second pedigree with autosomal dominant nemaline myopathy caused by TPM3 mutation: a clinical and pathological study. Neuromuscul Disord 2007; 17: 330–337.
Dufour C, Weinberger RP, Schevzov G, Jeffrey PL, Gunning P : Splicing of two internal and four carboxyl-terminal alternative exons in nonmuscle tropomyosin 5 pre-mRNA is independently regulated during development. J Biol Chem 1998; 273: 18547–18555.
Cooper JA : Actin dynamics: tropomyosin provides stability. Curr Biol 2002; 12: R523–R525.
Gordon AM, Homsher E, Regnier M : Regulation of contraction in striated muscle. Physiol Rev 2000; 80: 853–924.
Pittenger MF, Kazzaz JA, Helfman DM : Functional properties of non-muscle tropomyosin isoforms. Curr Opin Cell Biol 1994; 6: 96–104.
Dufour C, Weinberger RP, Gunning P : Tropomyosin isoform diversity and neuronal morphogenesis. Immunol Cell Biol 1998; 5: 424–429.
Cooley BC, Bergtrom G : Multiple combinations of alternatively spliced exons in rat tropomyosin-alpha gene mRNA: evidence for 20 new isoforms in adult tissues and cultured cells. Arch Biochem Biophys 2001; 390: 71–77.
Gunning PW, Schevzov G, Kee AJ, Hardeman EC : Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol 2005; 15: 333–341.
Durling HJ, Reilich P, Muller-Hocker J et al: De novo missense mutation in a constitutively expressed exon of the slow alpha-tropomyosin gene TPM3 associated with an atypical, sporadic case of nemaline myopathy. Neuromuscul Disord 2002; 12: 947–951.
Clarke N, Kolski H, Dye D et al: Mutations in TPM3 are a common cause of congenital fiber type disproportion. Ann Neurol (in press).
Sung SS, Brassington AM, Grannatt K et al: Mutations in genes encoding fast-twitch contractile proteins cause distal arthrogryposis syndromes. Am J Hum Genet 2003; 72: 681–690.
Lehtokari VL, Ceuterick-de Groote C, de Jonghe P et al: Cap disease caused by heterozygous deletion of the beta-tropomyosin gene TPM2. Neuromuscul Disord 2007; 17: 433–442.
Thierfelder L, Watkins H, MacRae C et al: Alpha-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell 1994; 77: 701–712.
Watkins H, Anan R, Coviello DA, Spirito P, Seidman JG, Seidman CE : A de novo mutation in alpha-tropomyosin that causes hypertrophic cardiomyopathy. Circulation 1995; 91: 2302–2305.
Karibe A, Tobacman LS, Strand J et al: Hypertrophic cardiomyopathy caused by a novel alpha-tropomyosin mutation (V95A) is associated with mild cardiac phenotype, abnormal calcium binding to troponin, abnormal myosin cycling, and poor prognosis. Circulation 2001; 103: 65–71.
Greenfield N, Huang Y, Swapna G et al: Solution NMR structure of the junction between tropomyosin molecules: implications for actin binding and regulation. J Mol Biol 2006; 364: 80–96.
Wang J, Sanger J, Kang S et al: Ectopic expression and dynamics of TPM1alpha and TPM1kappa in myofibrils of avian myotubes. Cell Motil Cytoskeleton 2007; 64: 767–776.
Jeannet PY, Mittaz L, Dunand M, Lobrinus JA, Bonafe L, Kuntzer T : Autosomal dominant nemaline myopathy: a new phenotype unlinked to previously known genetic loci. Neuromuscul Disord 2007; 17: 6–12.
Acknowledgements
We thank Drs B Hamel, H Kingston, M Lammens, C Müller-Reible, H Marschner-Schäfer, F Muntoni, Y Nevo, E Oberstzyn, S Reus and D Wenger for DNA samples from Turkish patients with nemaline myopathy in whom the mutation was not found. Marilotta Turunen and Dan Brudzewsky are thanked for technical assistance. Jaakko Sarparanta is acknowledged for kind help with figures.
Funding: VLL was supported by grants to CWP from the University of Helsinki, the Association Francaise contre les Myopathies, the Sigrid Jusélius Foundation, the Academy of Finland, the Finska Läkaresällskapet, and the Medicinska understödsföreningen Liv och Hälsa; KP was supported by grants from the University of Helsinki, the Association Francaise contre les Myopathies and the Oskar Öflund Foundation; AHB was supported by NIH AR44345, the Muscular Dystrophy Association of the USA, and the Joshua Frase Foundation. NGL was supported by Australian National Health and Medical Research Council (NH and MRC) Fellowship 403904.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lehtokari, VL., Pelin, K., Donner, K. et al. Identification of a founder mutation in TPM3 in nemaline myopathy patients of Turkish origin. Eur J Hum Genet 16, 1055–1061 (2008). https://doi.org/10.1038/ejhg.2008.60
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2008.60
Keywords
This article is cited by
-
Tropomyosin 3 (TPM3) function in skeletal muscle and in myopathy
Skeletal Muscle (2023)
-
Transcript-Based Diagnosis and Expanded Phenotype of an Intronic Mutation in TPM3 Myopathy
Molecular Diagnosis & Therapy (2022)
-
Nemaline myopathies: a current view
Journal of Muscle Research and Cell Motility (2019)
-
Chronic Megacolon Presenting in Adolescents or Adults: Clinical Manifestations, Diagnosis, and Genetic Associations
Digestive Diseases and Sciences (2019)
-
Congenital myopathies: disorders of excitation–contraction coupling and muscle contraction
Nature Reviews Neurology (2018)