Abstract
We have earlier described a syndrome characterized by microcephaly, cutis verticis gyrata, retinitis pigmentosa, cataracts, hearing loss and mental retardation (Mendelian inheritance in man (MIM) no: 605685) in two brothers from a non-consanguineous Lebanese family. In view of the rarity of the disorder and the high rate of inbreeding in the Lebanese population, we assumed an autosomal recessive trait inherited from a common ancestor. A genomewide scan was performed. The single locus on the long arm of chromosome 8 that showed homozygosity by descent comprised the gene responsible for Cohen syndrome (CS), VPS13B. We then sequenced VPS13B in the patients and found a homozygous splice site mutation. Several possible explanations for the overlap between CS and the clinical features observed in our patients are discussed. Our data highlight the potential of high-resolution homozygosity mapping in small populations with a high rate of inbreeding.
Similar content being viewed by others
Introduction
Mental retardation affects 1–3% of the general population,1 but only 3–7% can be recognized as defined syndromes. In as many as 30–50%, the cause is unknown.2
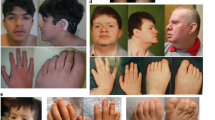
In 2001, we described a Lebanese family with syndromic mental retardation in two brothers. They exhibited microcephaly, primary cutis verticis gyrata of the scalp, prominent supraorbital ridges, coarse facial features (large nose, hypertelorism), retinitis pigmentosa, cataracts, sensorineural hearing loss and kyphoscoliosis in addition to mental retardation.3 As this combination of symptoms did not fit any known clinical entity, it was classified as a new syndrome (online Mendelian inheritance in man (OMIM) 605685) with either autosomal recessive or X-linked inheritance. Although parental consanguinity was not reported to be present in the family, we conducted a genomewide homozygosity mapping approach that identified a homozygous region on chromosome 8 in the patients. Within this candidate interval, we found a splice site mutation in VPS13B (OMIM 607817), the gene responsible for CS OMIM 216550) that results in a frameshift.
Methods
Patients
Clinical symptoms of the patients have been described earlier.3 DNA samples were available from the two affected brothers, one healthy brother, two healthy sisters and the mother (Figure 1a). RNA from whole blood was isolated using the PAXgene system (PreAnalytiX, Heidelberg, Germany). Written informed consent was obtained. The study was approved by the institutional review board of the Ethics Committee, University Hospital of Cologne.
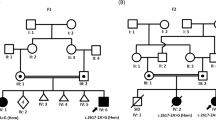
(a) Pedigree of the family investigated. (b) Graphical view of the LOD score calculation of genomewide SNP mapping under the assumption that the parents are unrelated (multiple peaks with low LOD values) and (c) that the parents are consanguineous. The resulting candidate locus on chromosome 8q22.1–q22.2 is indicated (arrow).
Linkage analysis
Assuming autosomal recessive inheritance, we performed genomewide homozygosity mapping using the Affymetrix GeneChip Human Mapping 10K Array, version 2.0 (Affymetrix, Santa Clara, CA, USA). GRR4 and PedCheck5 were used to verify relationships and to identify Mendelian errors. Non-parametric linkage analysis was carried out with MERLIN.6 Parametric linkage and haplotype analysis were performed using a modified version of the programs GENEHUNTER 2.17, 8 and ALLEGRO,9 assuming autosomal recessive inheritance. All data handling was performed using the graphical user interface ALOHOMORA.10 Graphic output of haplotypes was generated with HaploPainter.11
Mutation analysis
Direct sequencing of the 62 exons of VPS13B was conducted on genomic DNA. DNA numbering of mutations is based on the cDNA sequence of VPS13B (GenBank accession number: NM_017890). In all, 50 healthy control individuals (31 Lebanese and 19 Germans) were screened for the mutation identified. Reverse transcription from total RNA from whole blood was carried out with RevertAid™ H Minus M-MuLV Reverse Transcriptase (Fermentas), followed by cDNA-specific PCR amplification and direct sequencing using primers 5′-CACTGTGCACAAGTCAGTAG-3′ (forward, exon 50) and 5′-CATGATGAATTGAGCACTCG-3′ (reverse, exon 54). The connexin gene, GJB2, was also sequenced in one of the patients.
Results
If the parents are considered non-consanguineous, more than 20 chromosomal regions show segregation with the phenotype, each obtaining a combined maximum parametric logarithm of the odds (LOD) score of ∼1 (Figure 1b). In contrast, calculation under the hypothesis of parental consanguinity resulted in a single 7.24-Mb region on chromosome 8q22.1–q22.2, flanked by the single-nucleotide polymorphisms (SNPs) rs728185 and rs1954695, that shows homozygosity by descent (HBD) in both patients (LOD score of 2; Figure 1c). This region comprises 46 genes, including the gene for CS, VPS13B (earlier designated COH1). Molecular analysis of the complete coding region of VPS13B identified the c.9406-1G>C mutation in homozygous state (Figure 2b). This variant affects the invariant splice acceptor site of intron 51 in both patients. RT–PCR for the corresponding gene segment showed the activation of a cryptic acceptor splice site in exon 52, resulting in a 16-bp deletion in the mRNA (Figure 2d) predicted to lead to a frameshift and subsequent truncation of the protein to 3150 residues, p.Y3136TfsX16 (wild type: 4022 residues), or an unstable mRNA molecule that is rapidly degraded. The mutation was absent in 50 unrelated control individuals. Consistent with our mapping data, the mother and one sister are both heterozygous carriers of the mutation. We also screened the Connexin-26 gene, GJB2, the most prevalent cause of non-syndromic deafness, and did not identify any mutation.
(a) Heterozygous state for the c.9406–1G>C mutation in VPS13B as found in the mother and one sister (genomic sequence). The mutation affects the invariant acceptor splice site (AS) of intron 51. AS’ denotes the cryptic exonic splice site at position c.9420_9421 in exon 52. (b) Homozygosity for c.9406–1G>C in one of the index patients (genomic sequence). (c) RT–PCR analysis of VPS13B exons 50–54, wildtype. The 16-bp stretch deleted in the patients’ RNA (d) due to the c.9406–1G>C mutation and consecutive splicing at AS’ is boxed.
Discussion
We identified a novel VPS13B mutation in two brothers with symptoms compatible with CS, or at least Cohen-like syndrome, when applying the diagnostic criteria proposed by Chandler et al12 or Kolehmainen et al,13 respectively. Both present with two additional features, cutis verticis gyrata and sensorineural deafness, that have never been reported in CS patients. These unusual features may reflect a variant of CS, a syndrome known to be clinically highly heterogeneous. Also, several other explanations may underlie these additional features.
The mutation we identified, c.9406–1G>C, affects the same nucleotide as an earlier described mutation associated with CS, c.9406–1G>T, but leads to a different nucleotide change. Both nucleotides, C and T, at this position lead to the same abnormal splicing and the activation of a cryptic splice site near the 5′ end of exon 52 with a consecutive 16-bp frameshift deletion in the VPS13B mRNA.14 Interestingly, acceptor site prediction with NNSPLICE 0.9 (http://www.fruitfly.org/seq_tools/splice.html) for our mutation resulted in a much lower score for the cryptic AG (AG denotes the two nucleotides of the acceptor splice site) motif at c.9420_9421 (0.48) compared with c.9406–1G>T (0.71). However, we observed the same splicing pattern in total blood from our patients as was described for c.9406–1G>T.14 It is possible, but unlikely, that our mutation leads to a different splicing pattern or ratio between splice isoforms in other tissues that could explain the additional clinical features.
An alternative explanation for these features is an overlap of several genetic conditions. Consanguineous marriage represents a major risk for rare autosomal recessive diseases in offspring as it increases the frequency of homozygous genotypes. With an estimated 4–5 heterozygous recessive mutations per individual,15 offspring may become homozygous for mutations in unlinked genes – which can mimic a single syndromic disorder.16, 17 Thus, autosomal recessive hearing loss, as well as cutis verticis gyrata, could be caused by mutations in a different gene and locus. However, our linkage data do not support the presence of another causative homozygous locus. It is also possible that another mutated gene or modifier locus segregates in cis with the CS mutation and is responsible for deafness and cutis gyrata. On the basis of current knowledge, no deafness gene or locus has been mapped to the 8q22 homozygous region and the closest deafness gene, TFCP2L3, maps ∼1 Mb telomeric to its distal boundary. Given the high prevalence of hearing impairment and the multitude of genetic and environmental causes, this sensory deficit could well be because of variants or mutations in one or several additonal gene(s). Two out of the >20 chromosomal regions that segregate with the phenotype under the assumption of non-consanguinity contain known recessive deafness genes, CDH23 and MYO7A. As biallelic CDH23 or MYO7A mutations result in profound congenital deafness, and hearing impairment became evident in the third decade in our patients, a causative role of these genes is unlikely. The frequent association of cutis verticis gyrata with mental retardation18, 19 suggests that it may be a rare manifestation of CS. Consequently, VPS13B mutations should be considered if cutis verticis gyrata is observed in combination with mental retardation.
The homozygosity mapping approach conducted here can serve as a paradigm for linkage approaches in rare recessive phenotypes such as CS. For these diseases, the prevalence of homozygosity for the causative mutations can also be high in families without documented consanguinity, owing to a distant common ancestor. This is of particular relevance in small populations with a high rate of inbreeding.
Accession codes
References
Leonard H, Wen X : The epidemiology of mental retardation: challenges and opportunities in the new millennium. Ment Retard Dev Disabil Res Rev 2002; 8: 117–134.
Curry CJ, Stevenson RE, Aughton D et al: Evaluation of mental retardation: recommendations of a Consensus Conference: American College of Medical Genetics. Am J Med Genet 1997; 72: 468–477.
Megarbane A, Waked N, Chouery E et al: Microcephaly, cutis verticis gyrata of the scalp, retinitis pigmentosa, cataracts, sensorineural deafness, and mental retardation in two brothers. Am J Med Genet 2001; 98: 244–249.
Abecasis GR, Cherny SS, Cookson WO, Cardon LR : GRR: graphical representation of relationship errors. Bioinformatics 2001; 17: 742–743.
O’Connell JR, Weeks DE : PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 1998; 63: 259–266.
Abecasis GR, Cherny SS, Cookson WO, Cardon LR : Merlin – rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 2002; 30: 97–101.
Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES : Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 1996; 58: 1347–1363.
Strauch K, Fimmers R, Kurz T, Deichmann KA, Wienker TF, Baur MP : Parametric and nonparametric multipoint linkage analysis with imprinting and two-locus-trait models: application to mite sensitization. Am J Hum Genet 2000; 66: 1945–1957.
Gudbjartsson DF, Jonasson K, Frigge ML, Kong A : Allegro, a new computer program for multipoint linkage analysis. Nat Genet 2000; 25: 12–13.
Ruschendorf F, Nurnberg P : ALOHOMORA: a tool for linkage analysis using 10K SNP array data. Bioinformatics 2005; 21: 2123–2125.
Thiele H, Nurnberg P : HaploPainter: a tool for drawing pedigrees with complex haplotypes. Bioinformatics 2005; 21: 1730–1732.
Chandler KE, Kidd A, Al-Gazali L et al: Diagnostic criteria, clinical characteristics, and natural history of Cohen syndrome. J Med Genet 2003; 40: 233–241.
Kolehmainen J, Wilkinson R, Lehesjoki AE et al: Delineation of Cohen syndrome following a large-scale genotype-phenotype screen. Am J Hum Genet 2004; 75: 122–127.
Hennies HC, Rauch A, Seifert W et al: Allelic heterogeneity in the COH1 gene explains clinical variability in Cohen syndrome. Am J Hum Genet 2004; 75: 138–145.
Vogel F, Motulsky A : Human Genetics: Problems and Approaches, 3rd edn. New York: Springer-Verlag, 1997.
Ebermann I, Walger M, Scholl HP et al: Truncating mutation of the DFNB59 gene causes cochlear hearing impairment and central vestibular dysfunction. Hum Mutat 2007; 28: 571–577.
Ebermann I, Elsayed SM, Abdel-Ghaffar TY et al: Double homozygosity for mutations of AGL and SCN9A mimicking neurohepatopathy syndrome. Neurology 2008; 70: 2343–2344.
Dahir GA, Miller LK, Butler MG : Survey of mentally retarded males for cutis verticis gyrata and chromosomal fragile sites. Am J Med Genet 1992; 44: 118–120.
Larsen F, Birchall N : Cutis verticis gyrata: three cases with different aetiologies that demonstrate the classification system. Australas J Dermatol 2007; 48: 91–94.
Acknowledgements
We are indebted to the family that participated in this study. We thank Dr HC Hennies for kindly supplying VPS13B primers and Michaela Thoenes for technical assistance. This study was supported by the Pro Retina Deutschland, Pro-Re/KP/Bolz.1, and the Gertrud Kusen-Stiftung (HJB).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mégarbané, A., Slim, R., Nürnberg, G. et al. A novel VPS13B mutation in two brothers with Cohen syndrome, cutis verticis gyrata and sensorineural deafness. Eur J Hum Genet 17, 1076–1079 (2009). https://doi.org/10.1038/ejhg.2008.273
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2008.273