Abstract
We report on a patient with developmental delay and several facial characteristics reminiscent of Wolf–Hirschhorn syndrome, who carries a terminal 4p16.3 deletion of minimally 1.691 Mb and maximally 1.698 Mb. This deletion contains the FGFRL1 gene, but does not include the WHSC1 gene. Given its expression pattern and its involvement in bone and cartilage formation during embryonic development, the FGFRL1 gene represents a plausible candidate gene for part of the facial characteristics of Wolf–Hirshhorn syndrome in 4p16.3 deletion patients.
Similar content being viewed by others
Introduction
Wolf–Hirschhorn syndrome (WHS) is caused by variably sized deletions of chromosomal region 4p16.3.1 The core characteristics of WHS are pre- and post-natal growth delay, craniofacial dysgenesis, developmental delay and epilepsy.2 The severity of these characteristics is highly variable. Additional problems, including midline defects, occur at lower frequency in a subset of patients.
Typical craniofacial features include microcephaly, prominent glabella, widely spaced and prominent eyes, a ‘Greek warrior helmet appearance’ of the nose, a broad nasal tip, a short philtrum and downturned corners of the mouth.1 Identification of this condition strongly depends on recognition of the facial gestalt. Causative chromosomal deletions can be detected by regular karyotyping and smaller deletions can be readily demonstrated by fluorescence in situ hybridisation (FISH) with specific BAC probes.
Mapping of deletions in different WHS patients has allowed identification of two WHS critical regions (WHSCR and WHSCR-2), thought to be responsible for the core clinical manifestations of WHS.2, 3, 4 The WHSCR encompasses the WHSC2 gene and at least the 3′ portion of the WHSC1 gene (Figure 1).5 The WHSCR-2 contains the LETM1 gene as well as the 5′ portion of the WHSC1 gene.5 Deletion of the WHSC1 gene is supposed to be responsible for the facial appearance of WHS patients.2, 6 The present patient showed a 1.69 Mb deletion of the 4p16.3 region, not including the WHSC1 gene, despite the presence of some facial characteristics of WHS.
Materials and methods
Case report
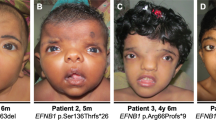
A 1.9-year-old girl presented with developmental delay. She was born at term as part of a dizygotic twin pregnancy. The pregnancy was characterized by intra-uterine growth retardation in the affected girl only, and breech position. Delivery was performed by caesarean section. Apgar scores were 9 and 10 at 1 and 5 min. Her birth weight was 2840 g (5th centile for age), and birth weight of her twin sister was 3130 g (16th centile for age). Her older brother and twin sister were healthy. The developmental delay was evident from the first months of life. She was hypotonic from birth. Her parents reported that she first laughed at 4 months. She sat at 11 months and could stand at the age of 19 months. No seizures were reported. Some language development was present from the age of 1.5 years. Clinical examinations revealed: body length of 0.81 m (5th centile for age), general hypotonia, hyperlaxity, and the following dysmorphic features: prominent glabella, large eyes, a nasal septum extending below the alae nasi, a very short philtrum and downturned corners of the mouth (Figure 2). These characteristics were absent in both parents and the healthy twin sister. Skull circumference was 49.2 cm (84th centile for age). The skull circumference at birth was not available, but at various ages the skull circumference was normal (84th centile for age). No hypertelorism was present; inner canthal distance: 2.8 cm (50th centile), outer canthal distance: 8.1 cm and interpupillary distance: 5.3 cm (both 50th – 75th centile). Mild developmental delay was determined with the Denver developmental scale. Metabolic investigations performed in urine and plasma showed results within the normal range. Additional investigations, including a renal ultrasound and an echocardiogram did not reveal any abnormalities.
Genetic investigations
In addition to prometaphase chromosome analysis with GTG banding at the 700-band level, FISH analysis with the GS-36-P21 probe, locating to the 4p subtelomeric region7 and a commercially available probe for WHS (LSI-WHS probe, Oncor, Gaithersburg, MD, USA) was performed because of facial characteristics reminiscent of WHS. The deletion was further characterized by a 4p tiling array CGH analysis, as previously described,8 and an Infinium HumanHap 300 Genotyping BeadChip SNP array analysis, using 750 ng of patient DNA, following the protocol as described by the manufacturer (Illumina Inc., San Diego, CA, USA). Dizygosity was confirmed by STR typing (AmpFISTR Profiler Plus, Perkin-Elmer Applied Biosystems, Foster City, CA, USA).
Results
The patient is one of dizygotic twins, with a normal female karyotype at regular chromosome analysis. A 4pter deletion was detected with the GS-36-P21 probe, but the LSI-WHS associated with common WHS deletions in band 4p16.3 was not deleted. In the parents no 4pter deletion was detected, demonstrating that the deletion had occurred de novo. Tiling BAC array CGH showed a deletion with the RP11-241P10 probe, but no deletion of the RP11-1170P16 probe (Figure 1). SNP array analysis to further narrow down the chromosome 4p16 break point showed all SNPs up to rs2854915 (nt:1.691.452) to be hemizygous, whereas from SNP rs2282763 (nt: 1.698.168) on, all SNPs gave diploid signals. Thus, the genome of our patient contained a terminal 4p16.3 deletion of minimally 1.691.452 bp and maximally 1.698.167 bp. This deletion is located telomeric to the WHSCR1 and WHSCR2 regions. The deletion break point region contains the final exon of the TMEM129 gene and the first 3 exons of the TACC3 gene (Figures 1 and 3). Twenty-six annotated genes locate telomeric to the TMEM gen.
Ensemble screen shot of chromosome 4p16. The SNP array analysis showed a terminal deletion of minimally 1.691.452 bp and maximally 1.698.167 bp. The minimal and maximal deletion are marked with the black lines. The red arrow demonstrates the FGFRL1 gene, a plausible candidate gene for part of the facial characteristics of Wolf–Hirschhorn syndrome in 4p16.3 deletion patients.
Discussion
Wolf–Hirschhorn syndrome is believed to represent a contiguous gene syndrome. Currently hemizygous deletion of the WHSC1 gene is considered to be the cause of the facial characteristics of WHS.2, 6 A recent description of two patients with deletions with break points near or within the distal boundary for WHSCR-2, thus excluding the WHSC1 gene, who presented with nonspecific manifestations, such as growth delay, developmental delay and feeding difficulties, but without facial characteristics of WHS, further corroborated this assumption.5
Here, we present a patient who raised clinical suspicion of WHS, although she did not show the typical ‘Greek helmet’ facial appearance. She had no microcephaly and only mild mental retardation. FISH analyses revealed a terminal 4p16.3 deletion that did not include the WHSC1 gene. This suggests that other genes, besides WHSC1, may contribute to features of the facial phenotype of WHS, such as a short philtrum and downturned corners of the mouth. Zollino et al.9 presented a patient with a very mild form of WHS, who carried no deletions as ascertained with R (RBG) banding and the available WHS probe D4S96 (Oncor). The relatively mild clinical manifestation of WHS, similar to our patient, could be consistent with a Pitt–Rogers–Danks (PRD) syndrome diagnosis. Possibly, more distal microdeletions are responsible for this milder facial WHS/PRD phenotype. Recent research using a 4p tiling BAC array CGH on 21 patients with WHS indicated a region ranging from 1.8 to 3 Mb from the p-telomere as the critical region for the typical WHS facial features.8 Our high resolution SNP array data indicate that some WHS facial features may map to the most distal 1.7 Mb of region 4p16.3, although we cannot exclude a position effect with certainty.
Searching for genes that in a hemizygous state may account for the observed facial anomalies in this patient, we consider the fibroblast growth factor receptor-like 1 (FGFRL1) to be a plausible candidate (Figure 3). This gene is situated centromeric of the approximately 0.6 Mb DNA, which can be deleted without an apparent phenotype.10 FGFRL1 is preferentially expressed in skeletal tissues and increases in its expression from a very low level after day 6 of mouse embryonic development.11, 12 The FGFRL1 gene shows particularly pronounced expression in the primordia of bones (eg, the maxillae, the mandibles, the ribs and the nose) and in the permanently cartilaginous tissues of the trachea. Although the muscle of the tongue and the diaphragm express FGFRL1 at a relatively high level, the heart, skeletal muscle, brain, lung, liver, kidney and gut express FGFRL1 at a basal level.11 The expression pattern of the FGFRL1 gene is consistent with the pattern of facial anomalies seen in our patient. Yet, it remains to be determined whether hemizygosity for the FGFRL1 gene may indeed account for some of the facial aspects of the WHS.
The deletion in this patient was not detected with the commercially available FISH probe for WHS (Oncor). These findings indicate that putative WHS patients should also be analysed with a more distal probe, especially those patients whose facial characteristics of WHS are accompanied by a relatively mild phenotype. Most importantly this case supports the notion that haploinsufficiency of genes distal to the WHSCR and WHSCR2 regions may contribute to the facial characteristics of WHS.
References
Titomanlio L, Romano A, Conti A et al: Mild Wolf–Hirschhorn phenotype and partial GH deficiency in a patient with a 4p terminal deletion. Am J Med Genet A 2004; 127: 197–200.
Bergemann AD, Cole F, Hirschhorn K : The etiology of Wolf–Hirschhorn syndrome. Trends Genet 2005; 21: 188–195.
Zollino M, Lecce R, Fischetto R et al: Mapping the Wolf–Hirschhorn syndrome phenotype outside the currently accepted WHS critical region and defining a new critical region, WHSCR-2. Am J Hum Genet 2003; 72: 590–597.
Rodriguez L, Zollino M, Climent S et al: The new Wolf–Hirschhorn syndrome critical region (WHSCR-2): a description of a second case. Am J Med Genet A 2005; 136: 175–178.
South ST, Bleyl SB, Carey JC et al: Two unique patients with novel microdeletions in 4p16.3 that exclude the WHS critical regions: implications for critical region designation. Am J Med Genet A 2007; 143: 2137–2142.
Van Buggenhout G, Melotte C, Dutta B et al: Mild Wolf–Hirschhorn syndrome: micro-array CGH analysis of atypical 4p16.3 deletions enables refinement of the genotype-phenotype map. J Med Genet 2004; 4: 691–698.
Knight SJ, Lese CM, Precht KS et al: An optimized set of human telomere clones for studying telomere integrity and architecture. Am J Hum Genet 2000; 67: 320–332.
Maas NM, Van Buggenhout G, Hannes F et al: Genotype-phenotype correlation in 21 patients with Wolf–Hirschhorn syndrome using high-resolution array CGH. J Med Genet 2008; 45: 71–80.
Zollino M, Bova R, Neri G : From Pitt–Rogers–Danks syndrome to Wolf–Hirschhorn syndrome and back? Am J Med Genet 1996; 66: 113–115.
Balikova I, Menten B, Ravel de G et al: Subtelomeric imbalances in phenotypically normal individuals. Hum Mutat 2007; 28: 958–967.
Trueb B, Taeschler S : Expression of FGFRL1, a novel fibroblast growth factor receptor, during embryonic development. Int J Mol Med 2006; 17: 617–620.
Trueb B, Zhuang L, Taeschler S, Wiedemann M : Characterization of FGFRL1, a novel fibroblast growth factor (FGF) receptor preferentially expressed in skeletal tissues. J Biol Chem 2003; 278: 33857–33865.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Engbers, H., van der Smagt, J., van ‘t Slot, R. et al. Wolf–Hirschhorn syndrome facial dysmorphic features in a patient with a terminal 4p16.3 deletion telomeric to the WHSCR and WHSCR 2 regions. Eur J Hum Genet 17, 129–132 (2009). https://doi.org/10.1038/ejhg.2008.168
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2008.168
Keywords
This article is cited by
-
From Wolf-Hirschhorn syndrome to NSD2 haploinsufficiency: a shifting paradigm through the description of a new case and a review of the literature
Italian Journal of Pediatrics (2022)
-
Distinct Epileptogenic Mechanisms Associated with Seizures in Wolf-Hirschhorn Syndrome
Molecular Neurobiology (2022)
-
Disorders of sex development in Wolf–Hirschhorn syndrome: a genotype–phenotype correlation and MSX1 as candidate gene
Molecular Cytogenetics (2021)
-
The first familial NSD2 cases with a novel variant in a Chinese father and daughter with atypical WHS facial features and a 7.5-year follow-up of growth hormone therapy
BMC Medical Genomics (2020)
-
Copy number variations associated with fetal congenital kidney malformations
Molecular Cytogenetics (2020)