Abstract
The presence or absence of genetic heterogeneity in Sicily has long been debated. Through the analysis of the variation of Y-chromosome lineages, using the combination of haplogroups and short tandem repeats from several areas of Sicily, we show that traces of genetic flows occurred in the island, due to ancient Greek colonization and to northern African contributions, are still visible on the basis of the distribution of some lineages. The genetic contribution of Greek chromosomes to the Sicilian gene pool is estimated to be about 37% whereas the contribution of North African populations is estimated to be around 6%.
In particular, the presence of a modal haplotype coming from the southern Balkan Peninsula and of its one-step derivates associated to E3b1a2-V13, supports a common genetic heritage between Sicilians and Greeks. The estimate of Time to Most Recent Common Ancestor is about 2380 years before present, which broadly agrees with the archaeological traces of the Greek classic era. The Eastern and Western part of Sicily appear to be significantly different by the χ2-analysis, although the extent of such differentiation is not very high according to an analysis of molecular variance. The presence of a high number of different haplogroups in the island makes its gene diversity to reach about 0.9. The general heterogeneous composition of haplogroups in our Sicilian data is similar to the patterns observed in other major islands of the Mediterranean, reflecting the complex histories of settlements in Sicily.
Similar content being viewed by others
Introduction
Sicily is the largest island in the Mediterranean basin and has served as a meeting place for different populations. Archaeological data indicate that this largest Mediterranean island was initially peopled by hunter-gathers approximately 10 000 years BP.1 Subsequent settlements may also have occurred prior to the transition to agriculture that began around 7000 years BP. During historical times, various ethnic groups that include Greeks, Phoenicians, Romans, Arabs and the Normans, left their legacy in Sicily.
Both the presence of genetic subdivision2, 3 and its absence4 have been reported in some analyses of classical polymorphism data. A more recent study using data from 9 autosomal microsatellite loci and 10 mitochondrial DNA (mtDNA) haplogroups (Hgs) reported a genetic differentiation consistent with that of other Mediterranean regions and correlated with longitude, although the causal underlying demography remained unclear.5
Several hypotheses have been proposed to explain this heterogeneity. It could be traced back to pre-Greek times when the Sicani, the first inhabitants of the island, were pushed westwards by the arrival of Siculi from Italy (about 1200 BP). Alternatively, the origin of such differentiation could be attributed to the Greek colonization between 2750 and 2200 BP in the southeastern region versus the west that was settled by Phoenicians (Figure 1a). Preliminary studies of the Y-chromosome Hg composition showed that approximately 60% of the Sicilian Y-chromosome Hgs are also prevalent in southern Italy and Greece.6 The presence of the lineage E3b1b-M81 in Sicily and Iberia reflects gene flows also from North Africa.7 But while Greek surnames display east–west differentiation,8, 9 the correlation of genetic diversification with longitude and the extent to which Greek colonization mediated gene-flow episodes remain still uncertain.
(a) Geographical map showing the main colonies by Greeks (triangles) and Phoenicians (circles) in the Mediterranean (seventh to sixth centuries BC). (b) Frequency distribution of the most representative haplotype 13-13-30-24-10-11-13 associated to the E3b1a2-V13 chromosomes in Sicily, in other populations taken from literature15, 23, 29, 30 and in samples from YHRD. The allelic combinations refer to the following order of loci: DYS19-DYS389I-DYS389II-DYS390-DYS391-DYS392-DYS393. (c) Frequency distribution of the haplotype 13-14-30-24-9-11-13 associated to the E3b1b-M81 chromosomes in Sicily (data from this study), in other populations taken from literature29 and in samples from YHRD.
Thanks to its haploid nature, Y-chromosome diversification is often highly correlated with geography.10 Recently, many authors showed that Y-chromosome combination of Hgs and short tandem repeats (STRs) are highly informative about the origin of male specific lineages, because of the detailed haplotypes that can be obtained and their geographical specificity.10, 11, 12, 13
In this paper, we evaluate the composition of Y-chromosome lineages using the combination of 33 biallelic markers and 12 STRs in samples coming from different areas of the island. We show diachronic genetic strata potentially linked to distinct historical colonization episodes within the Mediterranean basin. In addition, we estimate the extent of gene flow from both Greece and North Africa.
Materials and methods
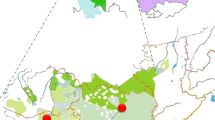
A total of 236 samples from 9 different areas of Sicily were studied. Latitudes (N) and longitudes (E) of each area are summarized in Figure 2.
(Top) The geographical map of the nine Sicilian samples is shown. Their latitude (N), longitude (E) and sample size are: (1) Trapani (TP) 38°07′, 12°07′, 33; (2) Mazara del Vallo (MZ) 37°65′, 12°58′, 18; (3) Santa Ninfa (SN) 37°77′, 12°88′, 31; (4) Alcamo (AL) 37°97′, 12°97′, 24; (5) Caccamo (CA) 37°93′, 13°07′, 16; (6) Sciacca (SC) 37°05′, 13°07′, 28; (7) Piazza Armerina (PZ) 37°38′, 14°37′, 28; (8) Troina (TR) 37°78′, 14°60′, 30; (9) Ragusa (RG) 36°93′, 14°75′, 28. The histogram plots the frequencies of the main haplogroups in the eastern and the western sides of the island.
Samples have further been grouped on the basis of historical and geographical criteria: western Sicily (WSI) includes 122 men from Trapani, Alcamo, Mazara del Vallo, Santa Ninfa and Caccamo; eastern Sicily (ESI) includes 114 men from Sciacca, Ragusa, Piazza Armerina and Troina. The partition reflects the history of the colonization occurred in the middle of the last millennium BC by the Greeks and the Phoenicians, who established their outposts in opposite parts of the island. We included Sciacca in the eastern part because of its ties to the important nearby Greek colony of Selinunte.
DNA extraction was carried out according to the phenol–chloroform protocol method. A set of 32 binary markers was tested, allowing us to assign the analysed Y chromosomes to Hgs. All polymorphisms have been previously reported.13, 14, 15, 16 The presence of the Y Alu polymorphic insertion was tested as described elsewhere.17 Genotyping was done by using the denaturing high performance liquid chromatography method proposed by Oefner and Underhill,18 with a phylogenetic hierarchical approach. The V12, V13, V22 polymorphisms, defining Hgs E3b1a1, E3b1a2 and E3b1a3, have been analysed as described in Cruciani et al.16 Data are referred to terminal mutation and according to the International Society of Genetic Genealogy nomenclature. The microsatellites DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393 and DYS385 A/B were analysed on all the samples. The E3b1a-M78 and the J2-M172 chromosomes were also typed for DYS439 plus DYS460 and DYS445 respectively. 5′-Fluorescently labelled PCR products were electrophoresed on an ABI PRISM 310 Genetic Analyzer. Genotypes were assigned by using control DNA samples provided by L Roewer (Institute of Legal Medicine, Charité, Berlin) and self-made allelic ladders provided by GeneScan software (Applied Biosystems). The DYS389II (AB fragment) allele number was determined by subtracting the DYS389I (CD fragment) repeat number.
Nomenclature is according to Kayser et al,19 with the exception of locus DYS389 where a monomorphic (TCTG)3 motif is included in the repeat count thus uniformly increasing the repeat size by three.
Gene diversity was calculated as described by Nei20 using the Arlequin software (http://cmpg.unibe.ch/software/arlequin3).
The proportion of genetic variance due to differences within or between populations was hierarchically apportioned through the analysis of molecular variance (AMOVA)21 included in the Arlequin software.
The admixture analysis code Admix2_0 was used to compute the estimators mY initially described in Bertorelle and Excoffier.22 The analysis was performed on E3b1b-M81 using as parental populations: Algerians and Tunisians (63 of 202 individuals), Egyptians (7 of 53 individuals) provided by Arredi et al23 and Continental Greeks (0 of 222 individuals; AP, unpublished data). A similar admixture analysis was performed for marker E3b1a2-V13 by using as parental populations: Greece (by pooling samples 30, 31 and 32, for a total of 35 out of 286 individuals) and northwestern Africa (by pooling samples from 35 up to 41, for a total of 2 out of 344 individuals) and northeastern Africa populations (by pooling samples from 43 up to 48, for a total of 3 out of 329 individuals) provided by Cruciani et al.12
The phylogenetic relationships were represented by the method of reduced median networks using the reduced median algorithm (r=2),24 followed by the median-joining algorithm (ɛ=0).25 The Network 4.1.1.2 software was used (www.fluxus-engineering.com).
The Time to Most Recent Common Ancestor (TMRCA) estimate and confidence interval (CI) were calculated using five STRs (DYS19, DYS391, DYS393, DYS439, DYS460) with the software Ytime v2.07 under the Simple Stepwise Mutation Model.26 The mutation rate used is the average of rates taken from Gusmão et al27 for DYS460 and from the Y Chromosome Haplotype Reference Database (YHRD, http://www.yhrd.org) for the other microsatellites.
Geographic maps built according to the Kriging procedure28 and implemented by the Surfer System (Golden Software) display STRs haplotype distributions under a particular Hg in Sicily and in other populations of the Mediterranean basin. Data were taken from the literature15, 23, 29, 30 and from YHRD. Principal component analysis was performed by using the R-package software v2.0.1 (http://www.r-project.org/).
Results and discussion
Haplogroups analysis
The 236 Y chromosomes are assigned to 24 different Hgs. Table 1 shows the Hg frequencies within the western and eastern regions as well as within each of the nine sampling locations.
Haplogroups common both to the European and Eurasian populations are present in Sicily. The most represented are R1b1c-M269 (24.58%), J2-M172 (15.25%) and E3b1a-M78 (11.44%). The co-occurrence of the Berber E3b1b-M81 (2.12%) and of the Mid-Eastern J1-M267 (3.81%) Hgs together with the presence of E3b1a1-V12, E3b1a3-V22, E3b1a4-V65 (5.5%) support the hypothesis of intrusion of North African genes.7, 12
Haplogroup R1b1c-M269, the most frequent Y-chromosome Hg in Europeans, is differentially distributed among eastern (18.4%) and western (30.3%) areas of Sicily. The Levantine Hgs7, 12 spread is also very informative: E3b1a-M78, G2-P15 and J2-M172 show frequencies (0.22, 0.32, 0.33), respectively. E3b1a2-V13 is present in both WSI (6.5%) and ESI (5.3%), whereas G2-P15 and J2-M172 are non-randomly distributed, occurring at higher frequencies in the eastern areas of the island (bar chart on the right of Figure 2 where E3b1a-M78 is reported without sub-Hgs).
Similarly, we could assign the 52% of the E3b1a-M78 chromosomes to E3b1a2-V13 that shows a clinal pattern of frequency distribution from the southern Balkan Peninsula (19.6%) to western Europe (2.5%)12 and is characterized by the otherwise rare nine-repeat allele at DYS460 locus.
In total, 48% of the E3b1a-M78 chromosomes in Sicily belongs to Hgs E3b1a1-V12, E3b1a3-V22 and E3b1a4-V65. These Hgs are common in northern Africa and are observed only in Mediterranean Europe12 and together the presence of the E3b1b-M81 highlights the genetic relationships between northern Africa and Sicily.
Furthermore Q-P36- or M242-derived chromosomes also detected significant similarities between Sicily (2.54%) and Lebanese populations (1.53%).30
The frequencies of the main Hgs (216 of 236 individuals) belonging to HgE, HgJ, HgI, HgG, R1b1c-M269 and R1a1-M17 are tested by the χ2-test that shows a highly significant difference between WSI and ESI (χ2=15.89, P=0.0072). The statistically significant P-value (P=0.009) of the genetic heterogeneity parameter Fst adds further evidence to such a genetic differentiation.
The AMOVA analysis indicates a variation among individuals within sampling areas and among sampling areas of 98.15 and 1.85%, respectively.
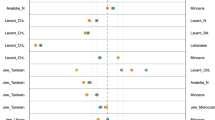
Principal component analysis (Figure 3) is performed on a database resulting from merging our present data with data from Pericic et al 200531 and with the more recent ones by Zalloua et al30 specifically devoted to Lebanon. Figure 3 displays a synthetic picture of the genetic distances between all the samples (WSI and ESI are compared with 28 other regions from Europe and the Mediterranean) according to the first and the second principal components of the frequencies of Hgs R1-M173, R1a1-M17, I1b-xM26-P37, E3b1a-M78 and J2b1-M102. The final result is that WSI and ESI are compared with 28 other regions from Europe and the Mediterranean. The general pattern in the graph is a separation between Balkan populations in the upper left corner (mainly due to I1b-xM26-P37), northern African and Mid-Eastern populations in the lower left corner and other European population on a branch extending to the right. ESI and WSI fall at the intersection of the latter clusters and are well separated from the Balkan populations. ESI is closer to Greece and Cyprus and it is discriminated by the first PC (66% total variance) from WSI (which is similar to Calabria and other western Europeans, mainly due to R1b1c-M269).
Plot of the two first principal coordinates (Principal Component Analysis, PCA). PCA is performed on a database resulting from merging our present data (ESI and WSI) with data from Pericic et al31 and with data from Zalloua et al30 regarding Lebanon. Population codes: AEI=Aegean islands; ALB=Albanian; ALG=Algerian (Arab); AND=Andalusian; BAS=Basque (French and Spanish); BEL=Belgian; BOS=Bosnian; CAT=Catalan; CRO=Croatian; CYP=Cypriot; DUT=Dutch; ESI=eastern Sicilian; FRE=French; GRE=Greek; HER=Herzegovinian; HUN=Hungarian; I-APU=Italian (Apulia); I-CAL=Italian (Calabria); I-SAR=Italian (Sardinia); LEB=Lebanese; MAL=Malta; MOR=Moroccan (Arab); ROM=Romanian; SER=Serbian; SLO=Slovenian; SPA=Spanish; TUN=Tunisian; TUR=Turkish (Istanbul); WSI=western Sicilian.
We assessed the relative contributions of the North African and Greek genes into the Sicily genetic pool by an admixture analysis, using Hg E3b1b-M81 for North Africa and E3b1a2-V13 for Greece. The estimated contribution resulted 6% for North Africa and 37.3% for Greece.
A high degree of gene diversity (0.904±0.011) has been calculated from this distribution of Hgs in Sicily (calculated using Table 1).
Microsatellite analysis
On the basis of the loci DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393 and DYS385 A/B, 158 different haplotypes are observed in Sicily. These loci form the core set included in the YHRD Database.32 Extended search in this database (which currently reports 52 655 haplotypes in a worldwide set of 464 populations) shows that the allelic frequencies in the Sicilian sample and Europeans do not differ significantly, with the exception of the DYS391 locus: in Sicily, the nine repeat allele occurs at a frequency of 11.7%, whereas it is very rare elsewhere in Europe; on the other hand it occurs in Tunisia at a frequency of 40%.23
The DYS445 microsatellite that normally shows an allele of 11–12 repeats has been observed with a deletion (only six repeats are present) in the J2 Hg. Such a deletion can be used as a stable unique event polymorphism defining a new subclade within J2a1, which has been named J2a1k.33 The DYS445-6 deletion is present in Greece (from 1.8 up to 3.5%) and in Crete (3.1%).29 Interestingly enough this DYS445-6 deletion is present in 46.67% of the J2 chromosomes in WSI against in 28.57% of the J2 chromosomes in ESI. In northern Italy about the 50% of the J2 chromosomes show the deletion, whereas in the southern Italy its frequency decreases (AP personal communication). Our data suggest that the presence of the DYS445-6 deletion is not due to the Greek colonization but it rather spread into the Mediterranean with the arrival of first farmers.
Lineage analysis
The combined evaluation of Hgs and STRs haplotypes provides clearer insights about the genetic structure of this island. Of the 181 different observed lineages 152 are unique, whereas 29 are observed more than once (Supplementary Table S1).
We have focused the analysis on the STRs haplotypes associated to E3b1a2-V13 and E3b1b-M81 chromosomes to investigate possible analogous backgrounds. Here we report on the STRs haplotypes with alleles taken from the following set of seven loci: DYS19-DYS389I-DYS389II-DYS390-DYS391-DYS392-DYS393.
Haplogroup E3b1a2-V13 shows a gene flows into Sicily from Greece and, more generally, from the southern Balkan Peninsula. The main haplotype related to this Hg, named southern Balkan Modal Haplotype, is defined by the allelic combination 13-13-30-24-10-11-13. This allelic combination belonging to V13 is common in Continental Greece 11.7%, Crete 3.62%29 and Albania (AP unpublished data).
In Figure 1b we analysed the distribution of ht 13-13-30-24-10-11-13 using data from this study data from15, 23, 29, 30 and data from YHRD.
It is also worth to note the presence of the Maghrebin haplotype 13-14-30-24-9-11-13 in Sicily (Figure 1c), which is associated to the E3b1b-M81 Hg, indicating a microsatellite background affinity with Tunisia where this lineage occurs at a frequency of 10%.23
The genetic relationships among the different populations involved in the history of the island are further explored by median-joining network analysis of the HgE using 5 STRs DYS19, DYS389I, DYS390, DYS391, DYS392. In Figure 4, the network subset that reports the chromosomes related to Hg E3b1a-M78 shows that Sicily and southeastern Europe, especially Greece and Albania, share a common background. Similarly, Hg E3b1b-M81 network cluster confirms the genetic affinity between Sicily and North Africa. These Y-chromosome lineages are consistent with the hypothesis that migrants from these different regions contributed to the genetic stratification of Sicily.
Network of haplogroup E. The microsatellites DYS19, DYS389I, DYS390, DYS391 and DYS392 were used. Areas of circles are proportional to the number of chromosomes (the smallest circle corresponds to two chromosomes). Areas of sectors are proportional to haplotype frequencies. Sources: Greece and Albania (AP unpublished data); North Africa;7, 23 southern Italy;7 Middle East.7, 15
Estimating TMRCA for E3b1 a2-V13 modal haplotype in the Mediterranean basin
The STRs haplotypes within Hg E3b1a2-V13 found in our Sicilian samples were merged with data from recent literature using the following microsatellites: DYS19, DYS391, DYS393, DYS439, DYS460.
We used populations from Cruciani et al12 (Sicilian, Continental Greek, Albanian, Greek from Crete, Greek from Aegean Islands, southwestern Turkish, southeastern Turkish, Turkish Cypriot, Central Anatolian) to obtain a homogeneous sample that could allow for tracing typical haplotypes and for dating TMRCA. Overall, we obtained 123 unrelated male subjects.
The 14 Sicilian samples out of these turned out to be tightly clustered around haplotype 13-10-13-12-9 and its one-step neighbours. Assuming the Sicilian Most Recent Common Ancestor had this modal haplotype and a male intergeneration time of 25 years, using the Ytime software we estimate a mean TMRCA of about 2380 years before present (CI: 675 to 6940 years).
Conclusions
As reviewed in the Introduction, Sicily has been subject to many different colonization episodes during pre-historical and historical times. The internal STR stratification of the E3b1a-M78 Hg reveals a certain amount of population structuring in the island, which is consistent with the successive presence of several different human populations. Specifically, some signatures of gene flows from Greece and from northern Africa can be identified.
The lineage E3b1a2-V13 13-13-30-24-10-11-13, which is typical of the Greek and southern Balkan regions, is present in the eastern side of the island and, together with the more general presence of the E3b1a2-V13 lineages, supports the presence of a common genetic heritage shared by the Sicilians and the Greeks. The TMRCA estimate of 2380 years BP, based on the STR lineages of the same E3b1a2-V13 Hg, coincides with the peak of the Greek classic era in Sicily, even if the very wide CI – a common result in such a kind of time estimates – however, we cannot exclude alternative hypotheses, for example an earlier arrivals of some of the E3b1a2-V13 chromosomes in Sicily from neolithic farmers.
We found a homogenous distribution of the E3b1a2-V13 marker over the island, which suggests an impact of the Greek colonization so impressive as to create a uniform stratum across Sicily. The Hg E3b1a2-V13 is estimated to contribute to the Sicilian gene pool by a fraction reaching 37%.
These data are compatible with the hypothesis that the largest historical demographic impact on Sicilian population was by the Greek settlers. A non-trivial question to raise for making this interpretation more plausible is whether the Greek colonies were of such size to lead to the diffusion of their genes. Because of the privileged position of Greece as ‘the door’ from the Near East to the Mediterranean, by the end of the Bronze Age the average density of the population was higher in Greece than in Europe by a factor of 3:3.7 inhabitants per square kilometre.34 Between 1000 and 400 BC, the population doubled in Europe, increasing from 10 to 20 million. In the same period the population trebled in Greece, reaching a total of 3 million. Around 400 BC Italy, the second most densely populated country in Europe after Greece, had about 4 million people.34 The Greek colonies of Sicily alone accounted for 1.5 million people, of which more than 10% (about 200 000) were of Greek origin.34, 35 To these Greek inhabitants of Sicily may be added at least another 100 000 Greek colonizers in southern Italy, so that before the Roman period, one in every 10–13 inhabitants in southern Italy was Greek.35 Admittedly these estimates must be taken with caution; however, their order of magnitude does not contradict the hypothesis of a possible introduction of Y lineages associated with Greek migration in ESI and in southern Italy.
The Hg E3b1b-M81, widely diffused in northwestern African populations, is estimated to contribute to the Sicilian gene pool at a rate of 6%. The distribution of E3b1b-M81 chromosomes in Africa closely matches the areas of distribution of Berber speaking populations, suggesting close Hg-ethnicity specificity. Interestingly, haplotype 13-14-30-24-9-11-13, associated to the E3b1b-M81 chromosomes,23, 36 is also present in Sicily. On the basis of the YHRD database, this haplotype occurs with high frequency in the Berber population of Tunisia, whereas it is less common elsewhere in North Africa.23, 37 The co-presence in Sicily of this haplotype and of the E3b1a1-V12, E3b1a3-V22, E3b1a4-V65 and J1-M267 Hgs could be attributed to the gene flows occurred during several trans-Mediterranean migrations from Africa, including the Arab invasion by sea.7, 12
The frequency of the R1b1c-M269 Hg, particularly high in some samples of WSI, is another interesting feature of the Sicilian paternal gene pool. This could be the legacy of the chromosomes coming from other parts of Europe. Moreover, Hgs I-M170, I1a-M253 and I1b2a-M223 are more represented in the northwestern area of the island than in the eastern area. Equally noteworthy is that the J2 chromosomes in WSI are also DYS445-6.
These differences lead us to a discussion of the genetic heterogeneity between areas of ESI and WSI. Such heterogeneity was previously emphasized on the study of the haemoglobinopathies,38 non-DNA polymorphisms,2 the autosomal microsatellites5 and surnames.8
In the present research, such heterogeneity is confirmed by the significantly different distribution of the frequencies of the main Hgs HgE, HgJ, HgI, HgG, R1b1c-M269 and R1a1-M17 and by the results from principal component analysis of such frequencies when compared to those from other parts of southern Europe and the Mediterranean, but weakly supported by a more sophisticated AMOVA analysis. This result is likely to be the effect of the contribution of different populations and repeated founder effects and it highlights the complex histories of settlement in this island.
The general heterogeneous composition of Hgs seen in our Sicilian data is consistent with similar patterns observed in other major islands of the Mediterranean, like Sardinia (gene diversity 0.801±0.010 SD on 939 samples using 23 Hgs)39 and Crete29, 40 (gene diversity 0.926±0.0006 SD on 193 samples using 29 Hgs),29 possibly reflecting the complex histories of settlements in these islands during the Holocene (Supplementary Table 1).
References
Malone C : The Italian Neolithic: a synthesis of research. J World Prehistory 2003; 17: 235–312.
Piazza A, Cappello N, Olivetti E, Rendine S : A genetic history of Italy. Ann Hum Genet 1988; 52: 203–213.
Cavalli Sforza LL, Menozzi P, Piazza A : The History and Geography of Human Genes. New Jersey: Princeton University, 1994.
Rickards O, Martinez-Labarga C, Scano G et al: Genetic history of the population of Sicily. Hum Biol 1998; 70: 699–714.
Romano V, Cali F, Ragalmuto A et al: Autosomal microsatellite and mtDNA genetic analysis in Sicily (Italy). Ann Hum Genet 2003; 67: 42–53.
Francalacci P, Morelli L, Underhill PA et al: Peopling of three Mediterranean islands (Corsica, Sardinia, and Sicily) inferred by Y-chromosome biallelic variability. Am J Phys Anthropol 2003; 121: 270–279.
Semino O, Magri C, Benuzzi G et al: Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am J Hum Genet 2004; 74: 1023–1034.
Guglielmino CR, Zei G, Cavalli-Sforza LL : Genetic and cultural transmission in Sicily as revealed by names and surnames. Hum Biol 1991; 63: 607–627.
De Silvestri A, Guglielmino CR : Sicilian provinces: population subdivisions revealed by surname frequencies. Hum Biol 2004; 76: 901–920.
Jobling MA, Tyler-Smith C : The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet 2003; 4: 598–612.
Moore LT, McEvoy B, Cape E, Simms K, Bradley DG : A Y-chromosome signature of hegemony in Gaelic Ireland. Am J Hum Genet 2006; 78: 334–338.
Cruciani F, La Fratta R, Trombetta B et al: Tracing past human male movements in northern/eastern Africa and western Eurasia: new clues from Y-chromosomal haplogroups E-M78 and J-M12. Mol Biol Evol 2007; 24: 1300–1311.
Underhill PA, Passarino G, Lin AA et al: The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 2001; 65: 43–62.
Y Chromosome Consortium: A Nomenclature System for the Tree of Human Y-chromosomal binary haplogroups. Genome Res 2002; 12: 339–348.
Cinnioglu C, King R, Kivisild T et al: Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet 2004; 114: 127–148.
Cruciani F, La Fratta R, Torroni A, Underhill PA, Scozzari R : Molecular dissection of the Y chromosome haplogroup E3B1A-M78 (E3b1a): a posteriori evaluation of a microsatellite-network-based approach through six new biallelic markers. Hum Mutat 2006; 27: 831–832.
Hammer MF, Horai S : Y chromosomal DNA variation and the peopling of Japan. Am J Hum Genet 1995; 56: 951–962.
Oefner PJ, Underhill PA : DNA mutation detection using denaturing high performance liquid chromatography (DHPLC); In: Dracopoli NC, Haines J, Korf BR, Morton C, Seidman CE, Seidman JG, Moir DT, Smith DR (eds). Current Protocols in Human Genetics, Suppl. 19 New York: Wiley, 1998, pp 7101–7112.
Kayser M, Krawczak M, Excoffier L et al: An extensive analysis of Y-chromosomal microsatellite haplotypes in globally dispersed human populations. Am J Hum Genet 2001; 68: 990–1018.
Nei M : Molecular Evolutionary Genetics. New York: Columbia University press, 1987.
Excoffier L, Smouse PE, Quattro JM : Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 1992; 131: 479–491.
Bertorelle G, Excoffier L : Inferring admixture proportions from molecular data. Mol Biol Evol 1998; 15: 1298–1311.
Arredi B, Poloni ES, Paracchini S, Zerjal T, Fathallah DM, Makrelouf A : Predominantly neolithic origin for Y-chromosomal DNA variation in North Africa. Am J Hum Genet 2004; 75: 338–345.
Bandelt HJ, Forster P, Röhl A : Median-joining networks for inferring intraspecific. Mol Biol Evol 1999; 16: 37–48.
Bandelt HJ, Forster P, Sykes BC, Richards MB : Mitochondrial portraits of human populations using median networks. Genetics 1995; 141: 743–753.
Behar DM, Thomas MG, Skorecki K et al: Multiple origins of Ashkenazi Levites: Y chromosome evidence for both Near Eastern and European ancestries. Am J Hum Genet 2003; 73: 768–779.
Gusmão L, Sánchez-Diz P, Calafell F et al: Mutation rates at Y chromosome specific microsatellites. Hum Mutat 2005; 26: 520–528.
Delfiner P : Linear estimation of non-stationary spatial phenomena; in Guarasio M, David M, Haijbegts C: advanced geostatistics in the mining industry. Dordrecht Reidel 1976; p: 49–68.
King RJ, Ozcan SS, Carter T et al: Differential Y-chromosome Anatolian influences on the Greek and Cretan Neolithic. Ann Hum Genet 2008; 72: 205–214.
Zalloua PA, Xue Y, Khalife J et al: Y-chromosomal diversity in Lebanon is structured by recent historical events. Am J Hum Genet 2008; 82: 873–882.
Pericic M, Lauc LB, Klaric IM et al: High-resolution phylogenetic analysis of Southeastern Europe traces major episodes of paternal gene flow among Slavic populations. Mol Biol Evol 2005; 22: 1964–1975.
Roewer L, Krawczak M, Willuweit S et al: Online reference database of European Y-chromosomal short tandem repeat (STR) haplotypes. Forensic Sci Int 2001; 118: 106–113.
Schrack BE, Athey TW, Wilson JF : Cluster analysis of extended Y-STR haplotypes leads to discovery of a large and widespread sub-clade of Y Haplogroup J2 pathway [abstract 994]. Presented at the Annual Meeting of The American Society of Human Genetics 2006, New Orleans, Louisiana.
McEvedy C, Jones R : Atlas of World Population History. Harmondsworth: Penguin Books Ltd, 1978.
Beloch J : Die Bevölkerung der griechisch-römischen Welt. Duncker and Humblot: Leipzig, 1886.
Robino C, Crobu F, Di Gaetano C et al: Analysis of Y-chromosomal SNP haplogroups and STR haplotypes in an Algerian population sample. Int J Legal Med 2008; 122: 251–255.
Luis JR, Rowold DW, Regueiro M et al: The Levantine versus the Horn of Africa: evidence for bi-directional corridors of human migrations. Am J Hum Genet 2004; 74: 532–544.
Schilirò G : Sicily: the world reservoir for thalassemias and haemoglobinopathies. Nature 1978; 276: 761–762.
Contu D, Morelli L, Santoni F, Foster JW, Francalacci P, Cucca F : Y-chromosome based evidence for pre-neolithic origin of the genetically homogeneous but diverse sardinian population: inference for association scans. PLoS ONE 2008; 3: e 1430.
Martinez L, Underhill PA, Zhivotovsky LA et al: Paleolithic Y-haplogroup heritage predominates in a Cretan highland plateau. Eur J Hum Genet 2007; 15: 485–493.
Acknowledgements
We are grateful to all blood donors for their generous participation in this study. Special thanks to Dr L Farina and Dr V Di Gaetano from AVIS provinciale Valderice who gave us a great help to collect some of these samples. We gratefully acknowledge Dr Alessandra Di Gregorio for her technical assistance. Very appropriate remarks from two anonymous referees contributed to a substantial revision of the paper and to a remarkable improvement of the data that is based on.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary information
Rights and permissions
About this article
Cite this article
Di Gaetano, C., Cerutti, N., Crobu, F. et al. Differential Greek and northern African migrations to Sicily are supported by genetic evidence from the Y chromosome. Eur J Hum Genet 17, 91–99 (2009). https://doi.org/10.1038/ejhg.2008.120
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2008.120
Keywords
This article is cited by
-
Characterizing the Mitochondrial Diversity of Arbi Goats from Tunisia
Biochemical Genetics (2021)
-
The spread of steppe and Iranian-related ancestry in the islands of the western Mediterranean
Nature Ecology & Evolution (2020)
-
A finely resolved phylogeny of Y chromosome Hg J illuminates the processes of Phoenician and Greek colonizations in the Mediterranean
Scientific Reports (2018)
-
Genetics of the peloponnesean populations and the theory of extinction of the medieval peloponnesean Greeks
European Journal of Human Genetics (2017)
-
The Italian genome reflects the history of Europe and the Mediterranean basin
European Journal of Human Genetics (2016)