Abstract
Northwest Siberia is geographically remote territory, which has been settled by indigenous human populations probably since the Upper Paleolithic. To investigate the genetic landscape of Northwest Siberians, we have analyzed mitochondrial and Y chromosome DNA polymorphisms of 169 unrelated individuals from Khanty and Mansi ethnic groups in Northwest Siberia. In addition, HVS-I sequences (N=3522) and Y chromosome SNP data (N=2175), obtained from the literature, were used to elucidate the genetic relationships among the North Eurasian populations. The results show clinal distributions of mtDNA and Y chromosome haplogroups along East–West axis of Northern Eurasia. In this context, the Ugric-speaking Khanty and Mansi appear as unique intermediate populations carrying Upper Paleolithic and more recent haplotypes typical for both West and East Eurasian gene pools. This admixture indicates that the Khanty and Mansi populations have resided in the contact zone of genetically distinguishable eastern and western Eurasia.
Similar content being viewed by others
Introduction
Anatomically modern humans populated the Siberian plain presumably during the Upper Paleolithic,1, 2 but the geographical origin of this population settlement is more controversial.3, 4 After the initial colonization, recurrent migrations from various directions have shaped the genetic composition of indigenous populations living in Siberia.5 For the current Siberian mtDNA and Y chromosome variation, both Central Asian6, 7, 8, 9 and East Asian origin has been proposed.9, 10, 11 In addition, South Siberia may have had an important role in the peopling of the more northern regions of Siberia.12, 13, 14, 15
One of the geographically remote and genetically less studied areas is Northwest Siberia, a region located north of the Central Asian Steppes, east from the Ural mountain range and surrounded by the Ob and Yenisei river valleys. In this study, we focus on the Northwest Siberian Ugric-speaking Khanty and Mansi populations. The Khanty and Mansi are known to have crossed the Urals, the northern geographical boundary between West and East Eurasia. They migrated from Pechora and Vychegda river regions west of Urals to the Ob river valley during the first millennium AD as one ancestral Ob-Ugric population, which split into the ethnic groups of Khanty and Mansi soon after this resettlement.5 The specific aim of the present study is to analyze mtDNA and Y chromosome variation in Khanty and Mansi populations, and situate them on the North Eurasian genetic landscape. Here North Eurasia is taken to include regions north of the 51° of northern latitude and from Northeast Europe to Northeast Siberia (Figure 1). Furthermore, we try to disentangle ancient migration processes from recent genetic amalgamation of West and East Eurasian gene pools in the Northwest of Siberia.
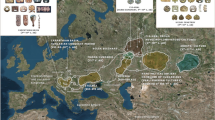
Map of the (a) 42 mtDNA and (b) 33 Y chromosome population samples analyzed with their approximate sampling locations. Circles are proportional to sample sizes and frequencies of geographically associated West Eurasian (mtDNA: HV, N1, N2, JT, UK, I, X; Y chromosome: F( × G, H, I, J, K), G, H, I, J, R1), East Eurasian (mtDNA: A, B, D, G, M7, M8-CZ, M9-E, M10, N9-Y, F; Y chromosome: C, K( × L, N, O, Q, R), L, N, Q, R( × R1) and South Asian (mtDNA: M*, R*; Y chromosome: O) haplogroup clusters are shown. Some subhaplogroups with distinct distribution in northern Eurasia are shown with individual colors (mtDNA U4, U7; Y chromosome N2, N3). Population abbreviations for mtDNA and Y chromosome and references therein can be found in Supplementary Tables 3 and 4, respectively.
Materials and methods
Khanty and Mansi samples
Blood samples with informed consents were collected from unrelated Khanty (n=106) and Mansi (n=63) individuals along the lower Ob-river valley. DNA was extracted following a standard phenol–chloroform protocol.
Mitochondrial HVS-I and II regions were amplified using primer pairs L15997-H16391 and L048-H408, respectively.16 Positions 16024–16383 and 72–340 were compared to the revised Cambridge Reference Sequence17 and variable sites were considered (data in Supplementary Table 1). Each mtDNA sequence was assigned to a haplogroup within the mtDNA phylogeny.12, 18, 19, 20, 21, 22 When coding region information was necessary to confirm haplogroup classification seven mtDNA positions (7028, 10400, 10873, 11151, 11719, 12308 and 12705)23 were analyzed (a subset of 41 samples; Table 1).
A set of 17 Y chromosome binary polymorphisms (SNPs) were hierarchically analyzed following the phylogeny and nomenclature of the Y chromosome24 in 28 Khanty and 25 Mansi samples. These 53 male samples were genotyped with markers M89, M172, M69, M201, M170, M9, 12f2, M145, M45, M173, SRY10831, M17, P25, M467, M1788, P437 and P367 as described in Bosch et al23 (additional primers in Supplementary Table 2) and their haplogroup was determined (Table 1). In addition, 11 Y chromosome short tandem repeat (STR) loci (DYS19, DYS390, DYS391, DYS392, DYS393, DYS385, DYS437, DYS438, DYS439, DYS389 I and II) were genotyped using the PowerplexY-kit (Promega, USA). In this data set, the actual repeat number at locus DYS389II was determined by substracting the length of DYS389I from DYS389II (STR haplotypes are available in Supplementary Table 2).
Reference data
A total of 3522 mitochondrial HVS-I sequences with relevant coding SNP/RFLP information from 29 different Eurasian populations were retrieved from the literature (Supplementary Table 3). Haplogroup classification was obtained from the original sources and refined according to the criteria used for the Khanty and Mansi samples. Thus, the S sequences15 were reclassified as M8a19 and the Saami HVS-I sequences25 were reclassified according to Bandelt et al.26 For the analysis, some sequences were clustered into major haplogroups: N1a and b into N1; W and N2a into N2; H and HV1 into HV as well as V into haplogroup HV0. Furthermore, sequences classified neither as M7, M8, M9, M10 and M13, nor as the major N or R haplogroups were considered to represent haplogroup M*. Two sequences were excluded: an L2 sequence in the Norwegian sample27 and one sequence with unclear haplogroup definition in the Karelian sample.25 The approximate locations of all the 42 mtDNA population samples used in this study are shown in Figure 1a.
Y chromosome SNP reference data for 2175 individuals from 27 different Eurasian populations were retrieved from the literature (Supplementary Table 4). All the analyzed individuals, typed with differing marker sets, were reclassified into 27 possible Y chromosome haplo/paragroups to allow population comparisons. The four Finnish individuals28 with no clear haplogroup classification were omitted and the Saami data8 was refined based on Tambets et al.29 The approximate locations of all the 33 Y chromosome population samples used in this study are shown in Figure 1b. In addition, nine-locus Y-STR minimal haplotype data of 1734 individuals from 10 different Eurasian populations were included in the STR comparisons (Supplementary Table 4).
Population samples were geographically grouped to Northeast Europe, Northwest Siberia, Northeast Siberia, Central Siberia, South Siberia, Southeast Siberia, Central Asia and East Asia based on Karafet et al7 (Figure 1a and b). Linguistic grouping of samples was performed according to Greenberg:30 Indo-European, Finnic, Ugric, Samoyedic, Turkic, Mongolic, Tunguso, Chukotko-Kamchatka, Eskimo-Aleut, Sino-Tibetan and language isolates of Yeniseian, Nivkhi and Yukagir (Supplementary Tables 3 and 4).
Data analysis
Population diversity indices for mtDNA and Y chromosomal data were estimated using Arlequin 3.01.31 In addition, the weighted intralineage mean pairwise difference (WIMP) was calculated.23, 32 The exact test for population differentiation and the analysis of molecular variance (AMOVA) were also performed.33 To define and test the geographic structure without a priori grouping, spatial analysis of molecular variance (SAMOVA)34 and autocorrelation indices for DNA analysis (AIDA)35 were performed.
To visualize population relationships, correspondence analyses (CA) were performed for both mtDNA and Y chromosome haplogroup frequencies using Statistica 6.0 (StatSoft Inc, US). Population pairwise FST (mtDNA) and RST (Y-STRs) distances were estimated and visualized by multidimensional scaling (MDS), computed with Statistica 6.0 (StatSoft Inc, US). The correlation between distance matrices of mtDNA and Y chromosome haplogroups was estimated using the Mantel test.31
Haplogroup-specific median joining networks36 for mitochondrial and Y chromosome data were constructed using program NETWORK 4.1 (www.fluxus-technology.com). Characters were weighted according to their variance within the haplogroup. For mitochondrial HVS-I positions, the weights given in Bandelt et al37 were used, with 99 assumed as weight for positions with no transitional information. For the Y-STR loci, the weights were calculated as 10 × (Vm/Vi), where Vm is the mean variance of all STR loci and Vi is the variance of STR locus in question.23 To estimate the age of expansion within the mtDNA haplogroups, the ρ-statistic implemented in Network 4.1 was used.38, 39
Results
Khanty and Mansi mtDNA and Y chromosome lineages
In the Khanty and Mansi population samples 39 and 27 haplotypes were observed which could be clustered into 19 and 13 mtDNA haplogroups, respectively (Table 1). These haplogroups represent both West and East Eurasian-associated mtDNA lineages (Figure 1a). Out of the 50 mtDNA haplotypes observed among the Khanty and Mansi, 16 haplotypes were observed in both populations (Supplementary Table 1). On the basis of mtDNA haplogroup frequencies, the Khanty and Mansi samples are not significantly different from each other, but they differ significantly (P<0.05) from the Mansi described by Derbeneva et al.40 However, based on mtDNA haplotype frequencies, the Khanty and two Mansi population samples in this study are significantly different from each other.
Y chromosome binary polymorphisms divided the Khanty into four and the Mansi into six paternal haplogroups (Table 1). Haplogroups R1a1, N2 and N3a were present in both populations, and comprised 96.4 and 84.0% of the Khanty and Mansi Y chromosomes, respectively. These Y chromosome haplogroups are typically found in West (R1a1)41 or across northern Eurasia (N2, N3a).11 On the basis of the Y chromosome haplogroup frequencies, the Khanty and Mansi are significantly different (P<0.05) from each other and also from the Khanty described by Karafet et al.7 The Khanty and Mansi Y-STR minimal haplotypes were compared to the YHRD database of 37 133 European (ie West Eurasia) and Asian (ie East Eurasia) haplotypes. This showed that 53.6% of the Khanty and 32.0% of the Mansi individuals carried haplotypes commonly found in West Eurasia, whereas 21.4% of the Khanty and 44.0% of the Mansi represent East Eurasian haplotype matches. When the binary SNPs were combined with the 11 Y-STR loci, the Khanty showed 14 and Mansi 13 distinct haplotypes with one common East Eurasian N211 haplotype (found in 17.9% of Khanty and 36.0% of Mansi) shared between these samples (Supplementary Table 2). However, the Khanty and Mansi are significantly different (P<0.05) from each other based on haplotype frequencies (Supplementary Table 2).
North Eurasian mtDNA and Y chromosome landscape
To provide a wider view of the North Eurasian genetic landscape, an mtDNA and Y chromosome dataset of 42 and 33 Eurasian population samples was compiled, respectively, (Supplementary Tables 3 and 4). Uniparental lineages were geographically classified into West Eurasian, East Eurasian and South Asian-associated haplogroups (Figure 1 legend).9, 11, 41, 42, 43 In this large dataset, both the mtDNA and Y chromosome data revealed similar patterns (Figure 1a and b). East Eurasian haplogroups are widespread across the whole Siberia, but show a clear decrease in frequency toward West Eurasia. In contrast, West Eurasian lineages show an opposite frequency trend, decreasing toward the East. AIDA showed significant clinal distribution of all mtDNA and Y chromosome haplogroups up to the distance of 4200 km (Figure 2). However, the Y chromosome haplogroup distribution showed a significant increase of autocorrelation at the distance of 4900 km, probably due to the high prevalence of N3 haplogroup across the whole northern Eurasia (Figure 1b). A Mantel test between the population pairwise distances of mtDNA and Y chromosome haplogroups among 15 Eurasian populations (see Supplementary Tables 3 and 4) showed nonsignificant correlation (P=0.41).
Genetic relationships among the North Eurasian populations
The MDS plot based on mtDNA haplotype FST distances (Figure 3a) places Khanty and Mansi samples in an intermediate position between Northeast Europe and South Siberia/Central Asia. The Y chromosome MDS plot (Figure 3b) appears less structured; here the Khanty and Mansi are only grouped in first dimension with the Finns. All the rest of the populations are loosely associated, with the exception of the distinct Buryat sample. CA of the mtDNA haplogroups shows a clinal west–east pattern of populations (Figure 4a), where the Khanty and Mansi are again located in an intermediate position between clusters of Northeast Europe and Central Asia/South Siberia. Compared to the mtDNA, the Y chromosome CA shows a more scattered pattern (Figure 4b), where the Northwest Siberians form their own cluster, with the exception of the distinct Ket and Selkup samples. The results of the SAMOVA analysis (not shown) are congruent with the MDS and CA plots (Figures 3a, b and 4a, b). The Khanty and Mansi group with the main cluster including populations from Northeast Europe, Central Asia and Central/South Siberia, but form their own cluster when the number of specified groups is increased.
Multidimensional scaling (MDS) based on (a) FST distances from HVS-I sequences (stress=0.115) and (b) RST distances from nine Y chromosome STR loci (stress=0.076) between 42 and 12 Eurasian population samples, respectively. Population abbreviations and reference can be found in Supplementary Tables 3 and 4.
Correspondence analysis based on (a) mtDNA and (b) Y chromosome haplogroups. Haplogroups are depicted and populations for mtDNA and Y chromosome are marked with numbers corresponding population samples and reference in Supplementary Tables 3 and 4, respectively.
The results of the AMOVA analyses showed a higher heterogeneity for the Y chromosome compared to the mtDNA (Table 2). When the populations are grouped according to geography or linguistics, the mtDNA and Y chromosome have similar distribution. However, the genetic structure of Y chromosomal haplogroups and haplotypes is better defined by geographical grouping (FCT; 10.61/14.61%) than by linguistic grouping (FCT; 7.35/13.80%), respectively.
mtDNA and Y chromosome diversity in Northwest Siberia
Out of all 383 Northwest Siberian mtDNA sequences in the present dataset (Supplementary Table 3), the most frequent West Eurasian haplogroups are U (27.2%), H (11.8%) and J (9.7%). Moreover, the frequency of the U4 mtDNA haplogroup among the Northwest Siberians is high, ranging from 8.5% among the Khanty to 28.9% among the Ket. The Northwest Siberian populations showed a high U4 haplotype diversity (0.833±0.024) compared to South Siberian (0.471±0.063) or Central Asian populations (0.400±0.237; Figure 5a). A similar pattern is detected within the U5a haplogroup with the highest diversity in Northeast Europe (0.930±0.015), followed by Northwest Siberia (0.713±0.083) and South Siberia (0.524±0.209). By contrast, the Northeast European U5b subhaplogroup, including the U5b1b1 ‘Saami motif’,25, 29 was not observed among Northwest Siberians except the Samoyedic-speaking Nganasan (1.9%). The U7 subhaplogroup was found only among the Northwest Siberian Khanty (14.2%) and Mansi (3.2–5.1%), Central Asian Uighur (4.2%) and Northeast European Finns (0.5%). The most frequent East Eurasian haplogroups in Northwest Siberia are C (19.1%) and D (15.7%), with C* and D* (up to 17.5 and 19.0% among the Mansi, respectively) the most frequent subhaplogroups. The C* subhaplogroup shows highest diversity in Central Asia (0.956±0.031) followed by South Siberia (0.893±0.011) and Northwest Siberia (0.869±0.021), similarly as haplogroup D* (data not shown).
Median-joining networks for Eurasian (a) mtDNA U4 sequences (colors for the U4 network are the following: gray (original Khanty and Mansi data with additional reference populations40, 44, 45); white (Northeast European populations16, 25, 46, 47, 48) and black (South Siberian populations12, 15, 49 grouped with Central Asian populations50, 51 and Central Siberia Yakuts52)) and Y chromosome (b) N3 (the N3 network corresponds the following colors: gray (original Khanty and Mansi data); white (Northeast European populations28, 53) and black (Central Siberian Yakuts54)) and (c) N2 Y-short tandem repeat (STR) haplotypes (nine loci) (the N2 network corresponds the following colors: gray (original Khanty and Mansi data); white (Northeast European populations11, 28) and black (South Siberian populations11 grouped with Northeast Siberian Eskimos11)).
Regarding the 587 Northwest Siberian Y chromosomes in the present dataset (see Supplementary Table 4), the most frequent haplogroups are N2 (33.4%), Q (23.7%) and N3 (22.3%). N2 and Q are at high frequency mainly within populations of Northwest Siberia (up to 92.1 and 93.8%, respectively), but N3 is frequent across the whole northern Eurasia (Figure 1b). Within haplogroup N3 all Khanty and Mansi STR haplotypes (Supplementary Table 2) cluster into the Northeast European clade as opposed to Central Siberian Yakut (Figure 5b). In contrast, within haplogroup N2, the Khanty and Mansi STR haplotypes (Supplementary Table 2) are divided between two previously defined11 West and East Eurasian clades (Figure 5c).
Discussion
The Khanty and Mansi possess a combination of east- and west-associated uniparental lineages, including features distinct from those observed among other Northwest Siberian populations. The distribution of the West and East Eurasian lineages shows clear east–west clines across the North Eurasia, which is in line with the current understanding of the Eurasian genetic landscape.8, 9, 11, 21, 22, 41 In this context, the Northwest Siberia appears as a ‘contact zone’ between the East and West Eurasian gene pools. Among the Northwest Siberians, the diversity within several uniparental sublineages (such as U7, J1b, J1c, J2, G2 and C5 for mtDNA; and N3 and R1a1 for Y chromosome) appears limited. However, c. 20–25% of the mtDNA haplotypes belonging to western (U4, U5a) and eastern (C*, D*) haplogroups show moderate haplotype diversities among the Khanty, Mansi, Ket and Nganasan. This suggests that the Northwest Siberia was initially colonized by humans carrying both West and East Eurasian Upper Paleolithic lineages. This is congruent with the concept of a genetic continuum of the early Upper Paleolithic populations expanding from Near East/Southeast Europe to Northwest Siberia.40, 44 Indeed, similar ρ-estimates were obtained for mtDNA U4 haplotypes in Northeast European (15.2–29.6 Kyr), Khanty and Mansi (11.9–29.7 Kyr) and all Northwest Siberian populations (12.8–29.6 Kyr), which also agree with the estimates presented for European U4 haplotypes (16–24 Kyr)18 In parallel, similar late Upper Paleolithic coalescent time estimates were observed for C* mtDNAs in Kanty and Mansi (14.5–31.7 Kyr BP) and all Northwest Siberian populations (14.4–32.0 Kyr BP).
However, the mtDNA U7 and Y chromosome N2 haplogroups among the Khanty and Mansi are probably of more recent origin. The U7 haplogroup is nearly absent across Northern Eurasia and mainly found in Near East (10–12% in Iran and India).9 All the U7 sequences found in Khanty and Mansi are identical, and the same sequence is found in Uighur of Central Asia. This suggests a recent founder effect of U7 haplogroup in Khanty and Mansi with a probable Central Asian origin. Similarly, the Southeast Asian-derived Y chromosome N2 lineage,11 allegedly specific for Uralic speakers, is also found among the Indo-European and Altaic speakers, but it is clearly more frequent in the Northwest Siberia (6.9–92.1%).7 Recently, two subclusters within the N2 lineage were described: the East (N2-A) and the West Eurasian (N2-E) clusters.11 Both East (N2-A) and West Eurasian (N2-E)-associated N2 subclusters are found among the Khanty and Mansi (Supplementary Table 2), whereas the Northeast European or South Siberian populations possess only either one of the subclusters (Figure 5c). The coalescent ages of these subclusters are considered relatively young (N2-E, 3.9–9.7; N2-A, 4.2–8.2 Kyr BP),11 although the diversity observed here within the N2-E subcluster (0.933±0.054) is significantly higher than in N2-A (0.500±0.121). This unique combination of N2 subclusters in the Khanty and Mansi suggests a recent amalgamation of western and eastern lineages in Northwest Siberia.
The haplogroup and haplotype compositions of the Khanty and Mansi samples differ significantly from those previously analyzed.7, 40 This, however, probably stems from limited sampling of different geographical subgroups as is probably in the case with the two Khanty population samples used in this study (T Karafet, personal communication) In addition, the histories of male and female populations appear to differ: Khanty and Mansi mtDNA exhibit between 58.7 and 68.9% of West Eurasian gene pool, whereas the other Northwest Siberians present 20.4–47.4% of western lineages. By contrast, the Khanty and Mansi Y chromosome shows between 76.6 and 89.3% of East Eurasian gene pool, similar values as estimated for the other Northwest Siberian populations.
The amalgamation of east- and west-associated uniparental lineages is also observed in Central Asia,50 Southwest Asia42 and South Siberia.14 However, the admixture among the Khanty and Mansi differs slightly from the surrounding populations. This is also supported by multilocus autosomal data, which clearly places the Khanty as an intermediate population between Northeast Europe and Central Siberia/East Asia.55
The initial admixture of uniparental lineages among the Khanty, Mansi and other Northwest Siberians could be explained by northward migration of human groups already carrying both West and East Eurasian Upper Paleolithic lineages originating from Central Asia and South Siberia. Later, when the Ob-ugric Khanty and Mansi migrated from the western side of Ural Mountains to Northwest Siberia, the unique amalgamation of N2-E and N2-A was formed. Similarly the mtDNA U7 and Y chromosome N3 haplotypes in Northwest Siberia suggest gene flow from Central Asia and Northeast Europe, respectively. However, Northwest Siberians are a heterogeneous group of populations showing lower haplotype diversity among and within uniparental haplogroups when compared to the more southern populations. This emphasize the complex background of Northwest Siberian genetic diversity, shaped by recurrent founder effects, admixture and drift in these indigenous populations.
References
Goebel T, Derevianko AP, Petrin VT : Dating the Middle-to-Upper-Paleolithic transition at Kara-Bom. Curr Anthropol 1993; 34: 452–458.
Vasil’ev SA, Kuzmin YV, Orlova LA, Dementiev VN : Radiocarbon-based chronology of the Paleolithic of Siberia and its relevance to the peopling of the New World. Radiocarbon 2002; 44: 503–530.
Dolukhanov PM, Shukurov AM, Tarasov PE, Zaitseva GI : Colonization of Northern Eurasia by modern humans: radiocarbon chronology and environment. J Archaeol Sci 2002; 29: 593–606.
Kuzmin YV, Keates SG : Comment on ‘colonization of Northern Eurasia by modern humans: radiocarbon chronology and environment’ by PM Dolukhanov, AM Shukurov, PE Tarasov and GI Zaitseva. Journal of Archaeological Science 29, 593–606 (2002). J Archaeol Sci 2004; 31: 141–143.
Kolga M, Tõnurist I, Vaba L, Viikberg J : The Red Book of the Peoples of the Russian Empire. Tallinn: NGO Red Book, 2001.
Karafet T, Xu L, Du R et al: Paternal population history of East Asia: sources, patterns, and microevolutionary processes. Am J Hum Genet 2001; 69: 615–628.
Karafet TM, Osipova LP, Gubina MA, Posukh OL, Zegura SL, Hammer MF : High levels of Y chromosome differentiation among native Siberian populations and the genetic signature of a boreal hunter-gatherer way of life. Hum Biol 2002; 74: 761–789.
Wells RS, Yuldasheva N, Ruzibakiev R et al: The Eurasian heartland: a continental perspective on Y chromosome diversity. Proc Natl Acad Sci USA 2001; 98: 10244–10249.
Metspalu M, Kivisild T, Metspalu E et al: Most of the extant mtDNA boundaries in south and southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans. BMC Genet 2004; 5: 26.
Su B, Xiao J, Underhill P et al: Y chromosome evidence for a northward migration of modern humans into Eastern Asia during the last Ice Age. Am J Hum Genet 1999; 65: 1718–1724.
Rootsi S, Zhivotovsky LA, Baldovic M et al: A counter-clockwise northern route of the Y chromosome haplogroup N from Southeast Asia towards Europe. Eur J Hum Genet 2007; 15: 204–211.
Derenko MV, Grzybowski T, Malyarchuk BA et al: Diversity of mitochondrial DNA lineages in South Siberia. Ann Hum Genet 2003; 67: 391–411.
Derenko M, Malyarchuk B, Denisova GA et al: Contrasting patterns of Y chromosome variation in South Siberian populations from Baikal and Altai-Sayan regions. Hum Genet 2006; 118: 591–604.
Derenko M, Malyarchuk B, Grzybowski T et al: Phylogeographic analysis of mitochondrial DNA in northern Asian populations. Am J Hum Genet 2007; 81: 1025–1041.
Starikovskaya EB, Sukernik RI, Derbeneva OA et al: Mitochondrial DNA diversity in indigenous populations of the southern extent of Siberia, and the origins of Native American haplogroups. Ann Hum Genet 2005; 69: 67–89.
Hedman M, Brandstätter A, Pimenoff V et al: Finnish mitochondrial DNA HVS-I and HVS-II population data. Forensic Sci Int 2007; 172: 171–178.
Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N : Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 1999; 23: 147.
Richards M, Macaulay V, Hickey E et al: Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet 2000; 67: 1251–1276.
Yao YG, Kong QP, Bandelt HJ, Kivisild T, Zhang YP : Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am J Hum Genet 2002; 70: 635–651.
Palanichamy MG, Sun C, Agrawal S et al: Phylogeny of mitochondrial DNA macrohaplogroup N in India, based on complete sequencing: implications for the peopling of South Asia. Am J Hum Genet 2004; 75: 966–978.
Tanaka M, Cabrera VM, Gonzalez AM et al: Mitochondrial genome variation in eastern Asia and the peopling of Japan. Genome Res 2004; 14: 1832–1850.
Torroni A, Achilli A, Macaulay V, Richards M, Bandelt HJ : Harvesting the fruit of the human mtDNA tree. Trends Genet 2006; 22: 339–345.
Bosch E, Calafell F, Gonzalez-Neira A et al: Paternal and maternal lineages in the Balkans show a homogeneous landscape over linguistic barriers, except for the isolated Aromuns. Ann Hum Genet 2006; 70: 459–487.
Y Chromosome Consortium: A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 2002; 12: 339–348.
Sajantila A, Lahermo P, Anttinen T et al: Genes and languages in Europe: an analysis of mitochondrial lineages. Genome Res 1995; 5: 42–52.
Bandelt HJ, Lahermo P, Richards M, Macaulay V : Detecting errors in mtDNA data by phylogenetic analysis. Int J Legal Med 2001; 115: 64–69.
Passarino G, Cavalleri GL, Lin AA, Cavalli-Sforza LL, Borresen-Dale AL, Underhill PA : Different genetic components in the Norwegian population revealed by the analysis of mtDNA and Y chromosome polymorphisms. Eur J Hum Genet 2002; 10: 521–529.
Lappalainen T, Koivumäki S, Salmela E et al: Regional differences among the Finns: a Y-chromosomal perspective. Gene 2006; 376: 207–215.
Tambets K, Rootsi S, Kivisild T et al: The western and eastern roots of the Saami – the story of genetic ‘outliers’ told by mitochondrial DNA and Y chromosomes. Am J Hum Genet 2004; 74: 661–682.
Greenberg JH : Indo-European and Its Closest Relatives: The Eurasiatic Language Family. Stanford: Stanford University Press, 2000.
Excoffier L, Laval G, Schneider S : Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 2005; 1: 47–50.
Hurles ME, Nicholson J, Bosch E, Renfrew C, Sykes BC, Jobling MA : Y chromosomal evidence for the origins of oceanic-speaking peoples. Genetics 2002; 160: 289–303.
Excoffier L, Smouse PE, Quattro JM : Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 1992; 131: 479–491.
Dupanloup I, Schneider S, Excoffier L : A simulated annealing approach to define the genetic structure of populations. Mol Ecol 2002; 11: 2571–2581.
Bertorelle G, Barbujani G : Analysis of DNA diversity by spatial autocorrelation. Genetics 1995; 140: 811–819.
Bandelt HJ, Forster P, Rohl A : Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 1999; 16: 37–48.
Bandelt HJ, Quintana-Murci L, Salas A, Macaulay V : The fingerprint of phantom mutations in mitochondrial DNA data. Am J Hum Genet 2002; 71: 1150–1160.
Forster P, Harding R, Torroni A, Bandelt HJ : Origin and evolution of Native American mtDNA variation: a reappraisal. Am J Hum Genet 1996; 59: 935–945.
Saillard J, Forster P, Lynnerup N, Bandelt HJ, Norby S : mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet 2000; 67: 718–726.
Derbeneva OA, Starikovskaya EB, Wallace DC, Sukernik RI : Traces of early Eurasians in the Mansi of northwest Siberia revealed by mitochondrial DNA analysis. Am J Hum Genet 2002; 70: 1009–1014.
Underhill PA, Shen P, Lin AA et al: Y chromosome sequence variation and the history of human populations. Nat Genet 2000; 26: 358–361.
Quintana-Murci L, Chaix R, Wells RS et al: Where west meets east: the complex mtDNA landscape of the southwest and Central Asian corridor. Am J Hum Genet 2004; 74: 827–845.
Shi H, Dong YL, Wen B et al: Y chromosome evidence of southern origin of the East Asian-specific haplogroup O3-M122. Am J Hum Genet 2005; 77: 408–419.
Derbeneva OA, Starikovskaia EB, Volod’ko NV, Wallace DC, Sukernik RI : Mitochondrial DNA variation in Kets and Nganasans and the early peoples of Northern Eurasia. Genetika 2002; 38: 1554–1560.
Goltsova TV, Osipova LP, Zhadanov SI, Villems R : The effect of marriage migration on the genetic structure of the Taimyr Nganasan population: genealogical analysis inferred from MtDNA markers. Genetika 2005; 41: 954–965.
Meinilä M, Finnilä S, Majamaa K : Evidence for mtDNA admixture between the Finns and the Saami. Hum Hered 2001; 52: 160–170.
Orekhov V, Poltoraus A, Zhivotovsky LA, Spitsyn V, Ivanov P, Yankovsky N : Mitochondrial DNA sequence diversity in Russians. FEBS Lett 1999; 445: 197–201.
Kornienko IV, Vodolazhskii DI, Afanas’eva GV, Ivanov PL : Polymorphism of the central region of D-loop of mitochondrial DNA and personality identification by forensic medicine methods. Sud Med Ekspert 2004; 47: 27–32.
Yao YG, Kong QP, Wang CY, Zhu CL, Zhang YP : Different matrilineal contributions to genetic structure of ethnic groups in the silk road region in china. Mol Biol Evol 2004; 21: 2265–2280.
Comas D, Plaza S, Wells RS et al: Admixture, migrations, and dispersals in Central Asia: evidence from maternal DNA lineages. Eur J Hum Genet 2004; 12: 495–504.
Comas D, Calafell F, Mateu E et al: Trading genes along the silk road: mtDNA sequences and the origin of central Asian populations. Am J Hum Genet 1998; 63: 1824–1838.
Pakendorf B, Wiebe V, Tarskaia LA et al: Mitochondrial DNA evidence for admixed origins of central Siberian populations. Am J Phys Anthropol 2003; 120: 211–224.
Kasperaviciute D, Kucinskas V, Stoneking M : Y chromosome and mitochondrial DNA variation in Lithuanians. Ann Hum Genet 2004; 68: 438–452.
Pakendorf B, Novgorodov IN, Osakovskij VL, Danilova AP, Protod’jakonov AP, Stoneking M : Investigating the effects of prehistoric migrations in Siberia: genetic variation and the origins of Yakuts. Hum Genet 2006; 120: 334–353.
Kidd KK, Pakstis AJ, Speed WC, Kidd JR : Understanding human DNA sequence variation. J Hered 2004; 95: 406–420.
Acknowledgements
We thank all the Khanty and Mansi donors for providing the blood samples. We thank Elena Bosch for the Y chromosome SNP-typing protocol, Kirsti Höök, Mònica Vallés, Stéphanie Plaza and Roger Anglada for technical support; T Karafet, L Rower, B Pakendorf and M Stoneking for the Eurasian Y chromosome reference data; Chiara Batini and Oscar Lao for help in the statistical analysis. Francesc Calafell and two anonymous referees are thanked for helpful comments on the paper. This research was financially supported by The Finnish Cultural Foundation, Federation of European Biochemical Societies and the Finnish Graduate School in Population Genetics (VNP), European contract grant (QLG2-CT-2001-00916), Direcció General de Recerca, Generalitat de Catalunya (2005SGR/00608, DC) and Academy of Finland (109265 and 111713, JUP; 80578, AS).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Rights and permissions
About this article
Cite this article
Pimenoff, V., Comas, D., Palo, J. et al. Northwest Siberian Khanty and Mansi in the junction of West and East Eurasian gene pools as revealed by uniparental markers. Eur J Hum Genet 16, 1254–1264 (2008). https://doi.org/10.1038/ejhg.2008.101
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2008.101
Keywords
This article is cited by
-
Genetic history of Bashkirian Mari and Southern Mansi ethnic groups in the Ural region
Molecular Genetics and Genomics (2019)
-
Complete human mtDNA genome sequences from Vietnam and the phylogeography of Mainland Southeast Asia
Scientific Reports (2018)
-
Genes reveal traces of common recent demographic history for most of the Uralic-speaking populations
Genome Biology (2018)
-
Analysis of ancient human mitochondrial DNA from the Xiaohe cemetery: insights into prehistoric population movements in the Tarim Basin, China
BMC Genetics (2015)
-
Y-SNP L1034: limited genetic link between Mansi and Hungarian-speaking populations
Molecular Genetics and Genomics (2015)